Zooming in on Transcription Preinitiation
- PMID: 27067110
- PMCID: PMC4906157
- DOI: 10.1016/j.jmb.2016.04.003
Zooming in on Transcription Preinitiation
Abstract
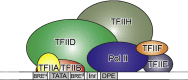
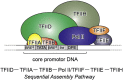
Class II gene transcription commences with the assembly of the Preinitiation Complex (PIC) from a plethora of proteins and protein assemblies in the nucleus, including the General Transcription Factors (GTFs), RNA polymerase II (RNA pol II), co-activators, co-repressors, and more. TFIID, a megadalton-sized multiprotein complex comprising 20 subunits, is among the first GTFs to bind the core promoter. TFIID assists in nucleating PIC formation, completed by binding of further factors in a highly regulated stepwise fashion. Recent results indicate that TFIID itself is built from distinct preformed submodules, which reside in the nucleus but also in the cytosol of cells. Here, we highlight recent insights in transcription factor assembly and the regulation of transcription preinitiation.
Keywords: RNA polymerase II; general transcription factor TFIID; multiprotein complex; preinitiation complex; transcription initiation.
Copyright © 2016 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
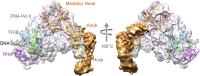




Similar articles
-
The general transcription machinery and general cofactors.Crit Rev Biochem Mol Biol. 2006 May-Jun;41(3):105-78. doi: 10.1080/10409230600648736. Crit Rev Biochem Mol Biol. 2006. PMID: 16858867 Review.
-
[Novel complex, formed by transcription coactivator SAYP].Mol Biol (Mosk). 2009 Nov-Dec;43(6):1055-62. Mol Biol (Mosk). 2009. PMID: 20088382 Russian.
-
Probing endogenous RNA polymerase II pre-initiation complexes by electrophoretic mobility shift assay.Methods Mol Biol. 2012;809:63-74. doi: 10.1007/978-1-61779-376-9_4. Methods Mol Biol. 2012. PMID: 22113268
-
Assembly of a mediator/TFIID/TFIIA complex bypasses the need for an activator.Curr Biol. 2003 Apr 29;13(9):772-7. doi: 10.1016/s0960-9822(03)00283-5. Curr Biol. 2003. PMID: 12725737
-
Regulation of the initiation of eukaryotic transcription.Essays Biochem. 2001;37:33-43. doi: 10.1042/bse0370033. Essays Biochem. 2001. PMID: 11758455 Review.
Cited by
-
Epigenetics and transcription regulation during eukaryotic diversification: the saga of TFIID.Genes Dev. 2019 Aug 1;33(15-16):888-902. doi: 10.1101/gad.300475.117. Epub 2019 May 23. Genes Dev. 2019. PMID: 31123066 Free PMC article.
-
Separable regulation of POW1 in grain size and leaf angle development in rice.Plant Biotechnol J. 2021 Dec;19(12):2517-2531. doi: 10.1111/pbi.13677. Epub 2021 Aug 14. Plant Biotechnol J. 2021. PMID: 34343399 Free PMC article.
-
Integration of transcription coregulator complexes with sequence-specific DNA-binding factor interactomes.Biochim Biophys Acta Gene Regul Mech. 2021 Oct;1864(10):194749. doi: 10.1016/j.bbagrm.2021.194749. Epub 2021 Aug 21. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 34425241 Free PMC article.
-
Insight into promoter clearance by RNA polymerase II.Proc Natl Acad Sci U S A. 2019 Nov 5;116(45):22426-22428. doi: 10.1073/pnas.1916582116. Epub 2019 Oct 18. Proc Natl Acad Sci U S A. 2019. PMID: 31628251 Free PMC article. No abstract available.
-
Mechanistic Differences in Transcription Initiation at TATA-Less and TATA-Containing Promoters.Mol Cell Biol. 2017 Dec 13;38(1):e00448-17. doi: 10.1128/MCB.00448-17. Print 2018 Jan 1. Mol Cell Biol. 2017. PMID: 29038161 Free PMC article.
References
-
- Kim Y.J., Bjorklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. - PubMed
-
- Thomas M.C., Chiang C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

