Colistin and Polymyxin B Dosage Regimens against Acinetobacter baumannii: Differences in Activity and the Emergence of Resistance
- PMID: 27067324
- PMCID: PMC4914682
- DOI: 10.1128/AAC.02927-15
Colistin and Polymyxin B Dosage Regimens against Acinetobacter baumannii: Differences in Activity and the Emergence of Resistance
Abstract
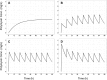
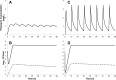
Infections caused by multidrug-resistant Acinetobacter baumannii are a major public health problem, and polymyxins are often the last line of therapy for recalcitrant infections by such isolates. The pharmacokinetics of the two clinically used polymyxins, polymyxin B and colistin, differ considerably, since colistin is administered as an inactive prodrug that undergoes slow conversion to colistin. However, the impact of these substantial pharmacokinetic differences on bacterial killing and resistance emergence is poorly understood. We assessed clinically relevant polymyxin B and colistin dosage regimens against one reference and three clinical A. baumannii strains in a dynamic one-compartment in vitro model. A new mechanism-based pharmacodynamic model was developed to describe and predict the drug concentrations and viable counts of the total and resistant populations. Rapid attainment of target concentrations was shown to be critical for polymyxin-induced bacterial killing. All polymyxin B regimens achieved peak concentrations of at least 1 mg/liter within 1 h and caused ≥4 log10 killing at 1 h. In contrast, the slow rise of colistin concentrations to 3 mg/liter over 48 h resulted in markedly reduced bacterial killing. A significant (4 to 6 log10 CFU/ml) amplification of resistant bacterial populations was common to all dosage regimens. The developed mechanism-based model explained the observed bacterial killing, regrowth, and resistance. The model also implicated adaptive polymyxin resistance as a key driver of bacterial regrowth and predicted the amplification of preexisting, highly polymyxin-resistant bacterial populations following polymyxin treatment. Antibiotic combination therapies seem the most promising option for minimizing the emergence of polymyxin resistance.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Polymyxin-resistant, carbapenem-resistant Acinetobacter baumannii is eradicated by a triple combination of agents that lack individual activity.J Antimicrob Chemother. 2017 May 1;72(5):1415-1420. doi: 10.1093/jac/dkx002. J Antimicrob Chemother. 2017. PMID: 28333347 Free PMC article.
-
Enhanced bacterial killing with colistin/sulbactam combination against carbapenem-resistant Acinetobacter baumannii.Int J Antimicrob Agents. 2021 Feb;57(2):106271. doi: 10.1016/j.ijantimicag.2020.106271. Epub 2020 Dec 23. Int J Antimicrob Agents. 2021. PMID: 33352235
-
An update on the arsenal for multidrug-resistant Acinetobacter infections: polymyxin antibiotics.Int J Infect Dis. 2015 Jan;30:125-32. doi: 10.1016/j.ijid.2014.10.014. Epub 2014 Nov 5. Int J Infect Dis. 2015. PMID: 25461655 Review.
-
In vitro pharmacodynamics of polymyxin B and tigecycline alone and in combination against carbapenem-resistant Acinetobacter baumannii.Antimicrob Agents Chemother. 2014;58(2):874-9. doi: 10.1128/AAC.01624-13. Epub 2013 Nov 25. Antimicrob Agents Chemother. 2014. PMID: 24277022 Free PMC article.
-
Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet?Int J Antimicrob Agents. 2016 Dec;48(6):592-597. doi: 10.1016/j.ijantimicag.2016.09.010. Epub 2016 Oct 18. Int J Antimicrob Agents. 2016. PMID: 27793510 Free PMC article. Review.
Cited by
-
Mechanism-Based Disease Progression Model Describing Host-Pathogen Interactions During the Pathogenesis of Acinetobacter baumannii Pneumonia.CPT Pharmacometrics Syst Pharmacol. 2018 Aug;7(8):507-516. doi: 10.1002/psp4.12312. Epub 2018 Aug 24. CPT Pharmacometrics Syst Pharmacol. 2018. PMID: 29761668 Free PMC article.
-
Emergence and Persistence of High-Risk Clones Among MDR and XDR A. baumannii at a Brazilian Teaching Hospital.Front Microbiol. 2019 Jan 4;9:2898. doi: 10.3389/fmicb.2018.02898. eCollection 2018. Front Microbiol. 2019. PMID: 30662431 Free PMC article.
-
Bacteremia by colistin-resistant Acinetobacter baumannii isolate: a case report.J Med Case Rep. 2019 May 8;13(1):141. doi: 10.1186/s13256-019-2062-3. J Med Case Rep. 2019. PMID: 31064407 Free PMC article.
-
Clinical Pharmacokinetics and Pharmacodynamics of Colistin.Clin Pharmacokinet. 2017 Dec;56(12):1441-1460. doi: 10.1007/s40262-017-0561-1. Clin Pharmacokinet. 2017. PMID: 28550595 Review.
-
In vitro Evaluation of the Colistin-Carbapenem Combination in Clinical Isolates of A. baumannii Using the Checkerboard, Etest, and Time-Kill Curve Techniques.Front Cell Infect Microbiol. 2017 May 24;7:209. doi: 10.3389/fcimb.2017.00209. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28596943 Free PMC article.
References
-
- Walker B, Barrett S, Polasky S, Galaz V, Folke C, Engstrom G, Ackerman F, Arrow K, Carpenter S, Chopra K, Daily G, Ehrlich P, Hughes T, Kautsky N, Levin S, Maler KG, Shogren J, Vincent J, Xepapadeas T, de Zeeuw A. 2009. Environment. Looming global-scale failures and missing institutions. Science 325:1345–1346. doi:10.1126/science.1175325. - DOI - PubMed
-
- Infectious Diseases Society of America (IDSA), Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, Schwartz D, Septimus E, Tenover FC, Gilbert DN. 2011. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis 52(Suppl 5):S397–S428. doi:10.1093/cid/cir153. - DOI - PMC - PubMed
-
- Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 55:3284–3294. doi:10.1128/AAC.01733-10. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

