Molar concentration from sequential 2-D water-window X-ray ptychography and X-ray fluorescence in hydrated cells
- PMID: 27067957
- PMCID: PMC4828672
- DOI: 10.1038/srep24280
Molar concentration from sequential 2-D water-window X-ray ptychography and X-ray fluorescence in hydrated cells
Abstract
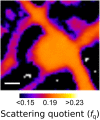
Recent developments in biological X-ray microscopy have allowed structural information and elemental distribution to be simultaneously obtained by combining X-ray ptychography and X-ray fluorescence microscopy. Experimentally, these methods can be performed simultaneously; however, the optimal conditions for each measurement may not be compatible. Here, we combine two distinct measurements of ultrastructure and elemental distribution, with each measurement performed under optimised conditions. By combining optimised ptychography and fluorescence information we are able to determine molar concentrations from two-dimensional images, allowing an investigation into the interactions between the environment sensing filopodia in fibroblasts and extracellular calcium. Furthermore, the biological ptychography results we present illustrate a point of maturity where the technique can be applied to solve significant problems in structural biology.
Figures




Similar articles
-
Simultaneous cryo X-ray ptychographic and fluorescence microscopy of green algae.Proc Natl Acad Sci U S A. 2015 Feb 24;112(8):2314-9. doi: 10.1073/pnas.1413003112. Epub 2015 Feb 9. Proc Natl Acad Sci U S A. 2015. PMID: 25675478 Free PMC article.
-
Mapping biological composition through quantitative phase and absorption X-ray ptychography.Sci Rep. 2014 Oct 28;4:6796. doi: 10.1038/srep06796. Sci Rep. 2014. PMID: 25348877 Free PMC article.
-
In Situ Ptychography of Heterogeneous Catalysts using Hard X-Rays: High Resolution Imaging at Ambient Pressure and Elevated Temperature.Microsc Microanal. 2016 Feb;22(1):178-88. doi: 10.1017/S1431927615015573. Microsc Microanal. 2016. PMID: 26914998
-
Nuclear microprobe: a microanalytical technique in biology.Cell Mol Biol (Noisy-le-grand). 1996 Feb;42(1):1-16. Cell Mol Biol (Noisy-le-grand). 1996. PMID: 8833662 Review.
-
Soft x-ray microscopy.Trends Cell Biol. 1999 Feb;9(2):44-7. doi: 10.1016/s0962-8924(98)01424-x. Trends Cell Biol. 1999. PMID: 10087616 Review.
Cited by
-
Correlative cellular ptychography with functionalized nanoparticles at the Fe L-edge.Sci Rep. 2017 Jul 6;7(1):4757. doi: 10.1038/s41598-017-04784-5. Sci Rep. 2017. PMID: 28684732 Free PMC article.
-
Resolving 500 nm axial separation by multi-slice X-ray ptychography.Acta Crystallogr A Found Adv. 2019 Mar 1;75(Pt 2):336-341. doi: 10.1107/S2053273318017229. Epub 2019 Feb 12. Acta Crystallogr A Found Adv. 2019. PMID: 30821266 Free PMC article.
-
High Reflectance Nanoscale V/Sc Multilayer for Soft X-ray Water Window Region.Sci Rep. 2017 Oct 10;7(1):12929. doi: 10.1038/s41598-017-13222-5. Sci Rep. 2017. PMID: 29018232 Free PMC article.
-
X-ray-Based Techniques to Study the Nano-Bio Interface.ACS Nano. 2021 Mar 23;15(3):3754-3807. doi: 10.1021/acsnano.0c09563. Epub 2021 Mar 2. ACS Nano. 2021. PMID: 33650433 Free PMC article.
-
Simultaneous structural and elemental nano-imaging of human brain tissue.Chem Sci. 2020 Aug 10;11(33):8919-8927. doi: 10.1039/d0sc02844d. Chem Sci. 2020. PMID: 34123146 Free PMC article.
References
-
- Mancuso A. P. et al. Internal structure of an intact Convallaria majalis pollen grain observed with X-ray Fresnel coherent diffractive imaging. Opt. Express 20, 26778–26785 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

