Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment to cis-regulatory modules
- PMID: 27068107
- PMCID: PMC4920159
- DOI: 10.1242/dev.131664
Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment to cis-regulatory modules
Abstract
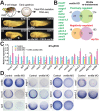
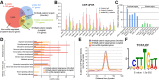
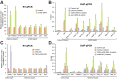
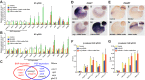
Key signalling pathways, such as canonical Wnt/β-catenin signalling, operate repeatedly to regulate tissue- and stage-specific transcriptional responses during development. Although recruitment of nuclear β-catenin to target genomic loci serves as the hallmark of canonical Wnt signalling, mechanisms controlling stage- or tissue-specific transcriptional responses remain elusive. Here, a direct comparison of genome-wide occupancy of β-catenin with a stage-matched Wnt-regulated transcriptome reveals that only a subset of β-catenin-bound genomic loci are transcriptionally regulated by Wnt signalling. We demonstrate that Wnt signalling regulates β-catenin binding to Wnt target genes not only when they are transcriptionally regulated, but also in contexts in which their transcription remains unaffected. The transcriptional response to Wnt signalling depends on additional mechanisms, such as BMP or FGF signalling for the particular genes we investigated, which do not influence β-catenin recruitment. Our findings suggest a more general paradigm for Wnt-regulated transcriptional mechanisms, which is relevant for tissue-specific functions of Wnt/β-catenin signalling in embryonic development but also for stem cell-mediated homeostasis and cancer. Chromatin association of β-catenin, even to functional Wnt-response elements, can no longer be considered a proxy for identifying transcriptionally Wnt-regulated genes. Context-dependent mechanisms are crucial for transcriptional activation of Wnt/β-catenin target genes subsequent to β-catenin recruitment. Our conclusions therefore also imply that Wnt-regulated β-catenin binding in one context can mark Wnt-regulated transcriptional target genes for different contexts.
Keywords: ChIP-seq; Gastrula; RNA-seq; Wnt signalling; Xenopus; β-catenin.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

References
-
- Akkers R. C., van Heeringen S. J., Jacobi U. G., Janssen-Megens E. M., Françoijs K.-J., Stunnenberg H. G. and Veenstra G. J. C. (2009). A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev. Cell 17, 425-434. 10.1016/j.devcel.2009.08.005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

