An eco-epidemiological study of Morbilli-related paramyxovirus infection in Madagascar bats reveals host-switching as the dominant macro-evolutionary mechanism
- PMID: 27068130
- PMCID: PMC4828640
- DOI: 10.1038/srep23752
An eco-epidemiological study of Morbilli-related paramyxovirus infection in Madagascar bats reveals host-switching as the dominant macro-evolutionary mechanism
Abstract
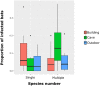
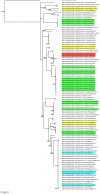
An eco-epidemiological investigation was carried out on Madagascar bat communities to better understand the evolutionary mechanisms and environmental factors that affect virus transmission among bat species in closely related members of the genus Morbillivirus, currently referred to as Unclassified Morbilli-related paramyxoviruses (UMRVs). A total of 947 bats were investigated originating from 52 capture sites (22 caves, 18 buildings, and 12 outdoor sites) distributed over different bioclimatic zones of the island. Using RT-PCR targeting the L-polymerase gene of the Paramyxoviridae family, we found that 10.5% of sampled bats were infected, representing six out of seven families and 15 out of 31 species analyzed. Univariate analysis indicates that both abiotic and biotic factors may promote viral infection. Using generalized linear modeling of UMRV infection overlaid on biotic and abiotic variables, we demonstrate that sympatric occurrence of bats is a major factor for virus transmission. Phylogenetic analyses revealed that all paramyxoviruses infecting Malagasy bats are UMRVs and showed little host specificity. Analyses using the maximum parsimony reconciliation tool CoRe-PA, indicate that host-switching, rather than co-speciation, is the dominant macro-evolutionary mechanism of UMRVs among Malagasy bats.
Figures



Similar articles
-
Highly diverse morbillivirus-related paramyxoviruses in wild fauna of the southwestern Indian Ocean Islands: evidence of exchange between introduced and endemic small mammals.J Virol. 2014 Aug;88(15):8268-77. doi: 10.1128/JVI.01211-14. Epub 2014 May 14. J Virol. 2014. PMID: 24829336 Free PMC article.
-
Paramyxoviruses from neotropical bats suggest a novel genus and nephrotropism.Infect Genet Evol. 2021 Nov;95:105041. doi: 10.1016/j.meegid.2021.105041. Epub 2021 Aug 17. Infect Genet Evol. 2021. PMID: 34411742
-
Evidence for retrovirus and paramyxovirus infection of multiple bat species in china.Viruses. 2014 May 16;6(5):2138-54. doi: 10.3390/v6052138. Viruses. 2014. PMID: 24841387 Free PMC article.
-
Paramyxoviruses in reptiles: a review.Vet Microbiol. 2013 Aug 30;165(3-4):200-13. doi: 10.1016/j.vetmic.2013.04.002. Epub 2013 Apr 9. Vet Microbiol. 2013. PMID: 23664183 Review.
-
Humanized animal viruses with special reference to the primate adaptation of morbillivirus.Vet Microbiol. 1992 Nov;33(1-4):275-86. doi: 10.1016/0378-1135(92)90055-x. Vet Microbiol. 1992. PMID: 1481360 Review.
Cited by
-
Preserve a Voucher Specimen! The Critical Need for Integrating Natural History Collections in Infectious Disease Studies.mBio. 2021 Jan 12;12(1):e02698-20. doi: 10.1128/mBio.02698-20. mBio. 2021. PMID: 33436435 Free PMC article. Review.
-
Zoonotic Paramyxoviruses: Evolution, Ecology, and Public Health Strategies in a Changing World.Viruses. 2024 Oct 29;16(11):1688. doi: 10.3390/v16111688. Viruses. 2024. Retraction in: Viruses. 2025 Jul 16;17(7):992. doi: 10.3390/v17070992. PMID: 39599803 Free PMC article. Retracted. Review.
-
Investigation of astrovirus, coronavirus and paramyxovirus co-infections in bats in the western Indian Ocean.Virol J. 2021 Oct 12;18(1):205. doi: 10.1186/s12985-021-01673-2. Virol J. 2021. PMID: 34641936 Free PMC article.
-
Diversity and seasonality of ectoparasite burden on two species of Madagascar fruit bat, Eidolon dupreanum and Rousettus madagascariensis.bioRxiv [Preprint]. 2025 Jan 22:2025.01.20.633693. doi: 10.1101/2025.01.20.633693. bioRxiv. 2025. Update in: Parasit Vectors. 2025 Jul 28;18(1):302. doi: 10.1186/s13071-025-06805-z. PMID: 39896656 Free PMC article. Updated. Preprint.
-
From Bat to Worse: The Pivotal Role of Bats for Viral Zoonosis.Microb Biotechnol. 2025 Jul;18(7):e70190. doi: 10.1111/1751-7915.70190. Microb Biotechnol. 2025. PMID: 40619741 Free PMC article. Review.
References
-
- Page R. D. M. Tangled Trees. Phylogeny, cospeciation and coevolution. Ethol ; 110, 577–578, doi: 10.1111/j.1439-0310.2004.00990.x (2003). - DOI
-
- Woolhouse M. E. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 10, S3–7 (2002). - PubMed
-
- Burland T. M. & Wilmer J. W. Seeing in the dark: molecular approaches to the study of bat populations. Biol. Rev. Camb. Philos. Soc. 76, 389–409 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

