ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles
- PMID: 27068479
- PMCID: PMC4828635
- DOI: 10.1038/srep23978
ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles
Abstract
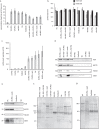
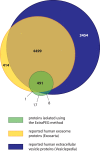
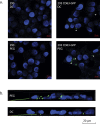
Initially thought to be a means for cells to eliminate waste, secreted extracellular vesicles, known as exosomes, are now understood to mediate numerous healthy and pathological processes. Though abundant in biological fluids, purifying exosomes has been challenging because their biophysical properties overlap with other secreted cell products. Easy-to-use commercial kits for harvesting exosomes are now widely used, but the relative low-purity and high-cost of the preparations restricts their utility. Here we describe a method for purifying exosomes and other extracellular vesicles by adapting methods for isolating viruses using polyethylene glycol. This technique, called ExtraPEG, enriches exosomes from large volumes of media rapidly and inexpensively using low-speed centrifugation, followed by a single small-volume ultracentrifugation purification step. Total protein and RNA harvested from vesicles is sufficient in quantity and quality for proteomics and sequencing analyses, demonstrating the utility of this method for biomarker discovery and diagnostics. Additionally, confocal microscopy studies suggest that the biological activity of vesicles is not impaired. The ExtraPEG method can be easily adapted to enrich for different vesicle populations, or as an efficient precursor to subsequent purification techniques, providing a means to harvest exosomes from many different biological fluids and for a wide variety of purposes.
Figures



References
-
- Trams E. G., Lauter C. J., Salem N. Jr. & Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochimica et biophysica acta 645, 63–70 (1981). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

