Horizontal gene transfer of acetyltransferases, invertases and chorismate mutases from different bacteria to diverse recipients
- PMID: 27068610
- PMCID: PMC4828791
- DOI: 10.1186/s12862-016-0651-y
Horizontal gene transfer of acetyltransferases, invertases and chorismate mutases from different bacteria to diverse recipients
Abstract
Background: Hoplolaimina plant-parasitic nematodes (PPN) are a lineage of animals with many documented cases of horizontal gene transfer (HGT). In a recent study, we reported on three likely HGT candidate genes in the soybean cyst nematode Heterodera glycines, all of which encode secreted candidate effectors with putative functions in the host plant. Hg-GLAND1 is a putative GCN5-related N-acetyltransferase (GNAT), Hg-GLAND13 is a putative invertase (INV), and Hg-GLAND16 is a putative chorismate mutase (CM), and blastp searches of the non-redundant database resulted in highest similarity to bacterial sequences. Here, we searched nematode and non-nematode sequence databases to identify all the nematodes possible that contain these three genes, and to formulate hypotheses about when they most likely appeared in the phylum Nematoda. We then performed phylogenetic analyses combined with model selection tests of alternative models of sequence evolution to determine whether these genes were horizontally acquired from bacteria.
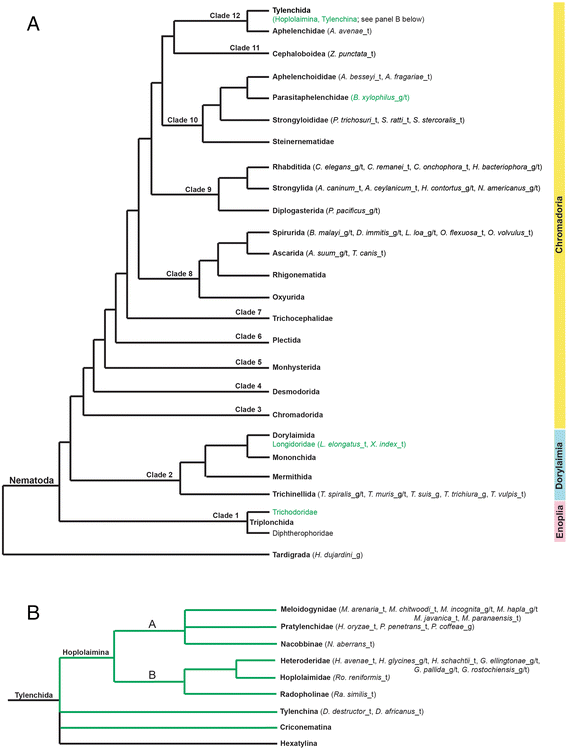
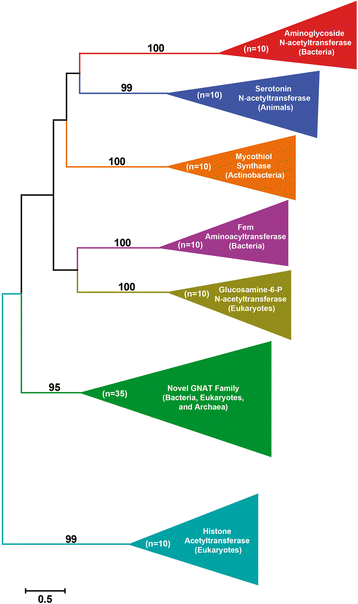
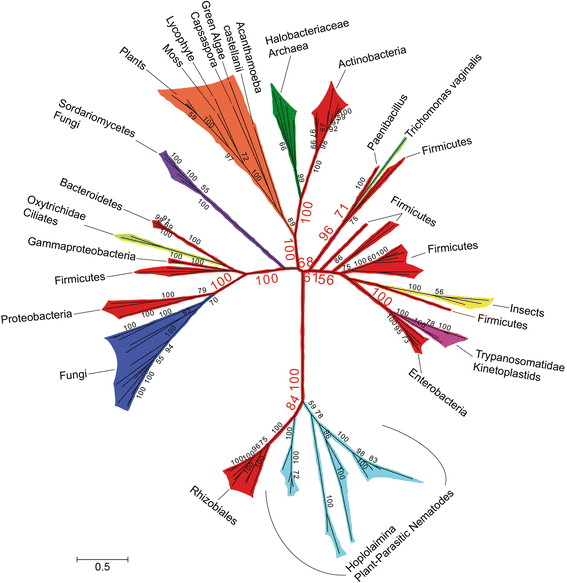
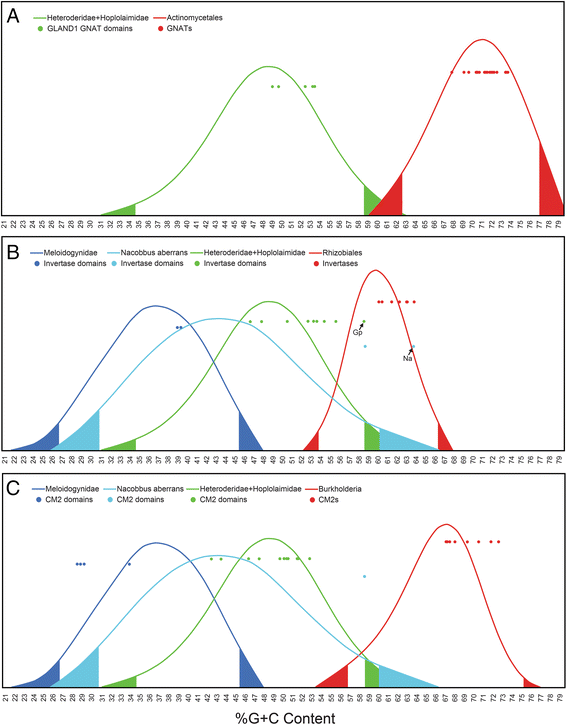
Results: Mining of nematode sequence databases determined that GNATs appeared in Hoplolaimina PPN late in evolution, while both INVs and CMs appeared before the radiation of the Hoplolaimina suborder. Also, Hoplolaimina GNATs, INVs and CMs formed well-supported clusters with different rhizosphere bacteria in the phylogenetic trees, and the model selection tests greatly supported models of HGT over descent via common ancestry. Surprisingly, the phylogenetic trees also revealed additional, well-supported clusters of bacterial GNATs, INVs and CMs with diverse eukaryotes and archaea. There were at least eleven and eight well-supported clusters of GNATs and INVs, respectively, from different bacteria with diverse eukaryotes and archaea. Though less frequent, CMs from different bacteria formed supported clusters with multiple different eukaryotes. Moreover, almost all individual clusters containing bacteria and eukaryotes or archaea contained species that inhabit very similar niches.
Conclusions: GNATs were horizontally acquired late in Hoplolaimina PPN evolution from bacteria most similar to the saprophytic and plant-pathogenic actinomycetes. INVs and CMs were horizontally acquired from bacteria most similar to rhizobacteria and Burkholderia soil bacteria, respectively, before the radiation of Hoplolaimina. Also, these three gene groups appear to have been frequent subjects of HGT from different bacteria to numerous, diverse lineages of eukaryotes and archaea, which suggests that these genes may confer important evolutionary advantages to many taxa. In the case of Hoplolaimina PPN, this advantage likely was an improved ability to parasitize plants.
Keywords: Evolution; Hoplolaimina; Horizontal gene transfer; Model selection analysis; Phylogenetics; Plant-parasitic nematodes.
Figures



Similar articles
-
Horizontal Gene Transfer from Bacteria Has Enabled the Plant-Parasitic Nematode Globodera pallida to Feed on Host-Derived Sucrose.Mol Biol Evol. 2016 Jun;33(6):1571-9. doi: 10.1093/molbev/msw041. Epub 2016 Feb 25. Mol Biol Evol. 2016. PMID: 26915958
-
Role of horizontal gene transfer in the evolution of plant parasitism among nematodes.Methods Mol Biol. 2009;532:517-35. doi: 10.1007/978-1-60327-853-9_30. Methods Mol Biol. 2009. PMID: 19271205 Review.
-
Selection of Heterodera glycines chorismate mutase-1 alleles on nematode-resistant soybean.Mol Plant Microbe Interact. 2005 Jun;18(6):593-601. doi: 10.1094/MPMI-18-0593. Mol Plant Microbe Interact. 2005. PMID: 15986929
-
Evolution of GHF5 endoglucanase gene structure in plant-parasitic nematodes: no evidence for an early domain shuffling event.BMC Evol Biol. 2008 Nov 3;8:305. doi: 10.1186/1471-2148-8-305. BMC Evol Biol. 2008. PMID: 18980666 Free PMC article.
-
Horizontal gene transfer in nematodes: a catalyst for plant parasitism?Mol Plant Microbe Interact. 2011 Aug;24(8):879-87. doi: 10.1094/MPMI-03-11-0055. Mol Plant Microbe Interact. 2011. PMID: 21539433 Review.
Cited by
-
Targeted transcriptomics reveals signatures of large-scale independent origins and concerted regulation of effector genes in Radopholus similis.PLoS Pathog. 2021 Nov 8;17(11):e1010036. doi: 10.1371/journal.ppat.1010036. eCollection 2021 Nov. PLoS Pathog. 2021. PMID: 34748609 Free PMC article.
-
Horizontal gene transfer from genetically modified plants - Regulatory considerations.Front Bioeng Biotechnol. 2022 Aug 31;10:971402. doi: 10.3389/fbioe.2022.971402. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36118580 Free PMC article.
-
Horizontal Gene Transfer Contributes to Plant Evolution: The Case of Agrobacterium T-DNAs.Front Plant Sci. 2017 Nov 24;8:2015. doi: 10.3389/fpls.2017.02015. eCollection 2017. Front Plant Sci. 2017. PMID: 29225610 Free PMC article. Review.
-
SCNBase: a genomics portal for the soybean cyst nematode (Heterodera glycines).Database (Oxford). 2019 Jan 1;2019(1):baz111. doi: 10.1093/database/baz111. Database (Oxford). 2019. PMID: 31680133 Free PMC article.
-
The genome of the soybean cyst nematode (Heterodera glycines) reveals complex patterns of duplications involved in the evolution of parasitism genes.BMC Genomics. 2019 Feb 7;20(1):119. doi: 10.1186/s12864-019-5485-8. BMC Genomics. 2019. PMID: 30732586 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

