ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy
- PMID: 27068792
- PMCID: PMC4828638
- DOI: 10.1038/srep24084
ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy
Abstract
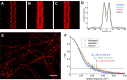
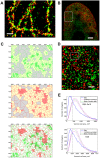
Super-resolution microscopy (PALM, STORM etc.) provides a plethora of fluorescent signals in dense cellular environments which can be difficult to interpret. Here we describe ClusterViSu, a method for image reconstruction, visualization and quantification of labelled protein clusters, based on Voronoi tessellation of the individual fluorescence events. The general applicability of this clustering approach for the segmentation of super-resolution microscopy data, including for co-localization, is illustrated on a series of important biological objects such as chromatin complexes, RNA polymerase, nuclear pore complexes and microtubules.
Figures


References
-
- Szymborska A. et al. Nuclear Pore Scaffold Structure Analyzed by Super-Resolution Microscopy and Particle Averaging. Science 341, 655–658 (2013). - PubMed
-
- Banterle N., Khanh H. B., Lemke E. A. & Beck M. Fourier ring correlation as a resolution criterion for super-resolution microscopy. J. Struct. Biol. 183, 363–367 (2013). - PubMed
-
- Baddeley D., Cannell M. B. & Soeller C. Visualization of Localization Microscopy Data. Microsc. Microanal. 16, 64–72 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

