Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
- PMID: 27069501
- PMCID: PMC4812495
- DOI: 10.1155/2016/8356294
Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
Abstract
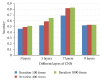
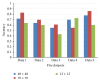
Early brain tumor detection and diagnosis are critical to clinics. Thus segmentation of focused tumor area needs to be accurate, efficient, and robust. In this paper, we propose an automatic brain tumor segmentation method based on Convolutional Neural Networks (CNNs). Traditional CNNs focus only on local features and ignore global region features, which are both important for pixel classification and recognition. Besides, brain tumor can appear in any place of the brain and be any size and shape in patients. We design a three-stream framework named as multiscale CNNs which could automatically detect the optimum top-three scales of the image sizes and combine information from different scales of the regions around that pixel. Datasets provided by Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) organized by MICCAI 2013 are utilized for both training and testing. The designed multiscale CNNs framework also combines multimodal features from T1, T1-enhanced, T2, and FLAIR MRI images. By comparison with traditional CNNs and the best two methods in BRATS 2012 and 2013, our framework shows advances in brain tumor segmentation accuracy and robustness.
Figures
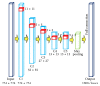
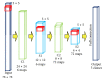

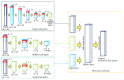
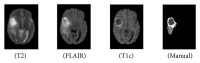


References
-
- Havaei M., Davy A., Warde-Farley D., et al. Brain tumor segmentation with deep neural networks. http://arxiv.org/abs/1505.03540v1. - PubMed
-
- Jiang J., Wu Y., Huang M. Y., Yang W., Chen W., Feng Q. 3D brain tumor segmentation in multimodal MR images based on learning population- and patient-specific feature sets. Computerized Medical Imaging and Graphics. 2013;37(7):512–521. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

