ToxR Antagonizes H-NS Regulation of Horizontally Acquired Genes to Drive Host Colonization
- PMID: 27070545
- PMCID: PMC4829181
- DOI: 10.1371/journal.ppat.1005570
ToxR Antagonizes H-NS Regulation of Horizontally Acquired Genes to Drive Host Colonization
Abstract
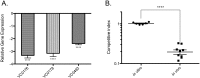
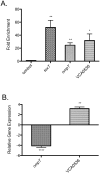
The virulence regulator ToxR initiates and coordinates gene expression needed by Vibrio cholerae to colonize the small intestine and cause disease. Despite its prominence in V. cholerae virulence, our understanding of the direct ToxR regulon is limited to four genes: toxT, ompT, ompU and ctxA. Here, we determine ToxR's genome-wide DNA-binding profile and demonstrate that ToxR is a global regulator of both progenitor genome-encoded genes and horizontally acquired islands that encode V. cholerae's major virulence factors and define pandemic lineages. We show that ToxR shares more than a third of its regulon with the histone-like nucleoid structuring protein H-NS, and antagonizes H-NS binding at shared binding locations. Importantly, we demonstrate that this regulatory interaction is the critical function of ToxR in V. cholerae colonization and biofilm formation. In the absence of H-NS, ToxR is no longer required for V. cholerae to colonize the infant mouse intestine or for robust biofilm formation. We further illustrate a dramatic difference in regulatory scope between ToxR and other prominent virulence regulators, despite similar predicted requirements for DNA binding. Our results suggest that factors in addition to primary DNA structure influence the ability of ToxR to recognize its target promoters.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Stringent response interacts with the ToxR regulon to regulate Vibrio cholerae virulence factor expression.Arch Microbiol. 2020 Aug;202(6):1359-1368. doi: 10.1007/s00203-020-01847-6. Epub 2020 Mar 10. Arch Microbiol. 2020. PMID: 32157346
-
Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization.Proc Natl Acad Sci U S A. 2000 Aug 29;97(18):10220-4. doi: 10.1073/pnas.170219997. Proc Natl Acad Sci U S A. 2000. PMID: 10944196 Free PMC article.
-
Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro).mBio. 2013 Aug 27;4(5):e00366-13. doi: 10.1128/mBio.00366-13. mBio. 2013. PMID: 23982069 Free PMC article.
-
Expression of Vibrio cholerae virulence genes in response to environmental signals.Curr Issues Intest Microbiol. 2002 Sep;3(2):29-38. Curr Issues Intest Microbiol. 2002. PMID: 12400636 Review.
-
Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae.Mol Microbiol. 1992 Feb;6(4):451-8. doi: 10.1111/j.1365-2958.1992.tb01489.x. Mol Microbiol. 1992. PMID: 1560773 Review.
Cited by
-
Characterization of MxiE- and H-NS-Dependent Expression of ipaH7.8, ospC1, yccE, and yfdF in Shigella flexneri.mSphere. 2022 Dec 21;7(6):e0048522. doi: 10.1128/msphere.00485-22. Epub 2022 Nov 8. mSphere. 2022. PMID: 36346241 Free PMC article.
-
Ambient pH Alters the Protein Content of Outer Membrane Vesicles, Driving Host Development in a Beneficial Symbiosis.J Bacteriol. 2019 Sep 20;201(20):e00319-19. doi: 10.1128/JB.00319-19. Print 2019 Oct 15. J Bacteriol. 2019. PMID: 31331976 Free PMC article.
-
Vibrio cholerae Virulence Activator ToxR Regulates Manganese Transport and Resistance to Reactive Oxygen Species.Infect Immun. 2020 Feb 20;88(3):e00944-19. doi: 10.1128/IAI.00944-19. Print 2020 Feb 20. Infect Immun. 2020. PMID: 31871097 Free PMC article.
-
Antibiotic resistance and virulence genes profiling of Vibrio cholerae and Vibrio mimicus isolates from some seafood collected at the aquatic environment and wet markets in Eastern Cape Province, South Africa.PLoS One. 2023 Aug 24;18(8):e0290356. doi: 10.1371/journal.pone.0290356. eCollection 2023. PLoS One. 2023. PMID: 37616193 Free PMC article.
-
H-NS-like proteins in Pseudomonas aeruginosa coordinately silence intragenic transcription.Mol Microbiol. 2021 Jun;115(6):1138-1151. doi: 10.1111/mmi.14656. Epub 2020 Dec 18. Mol Microbiol. 2021. PMID: 33245158 Free PMC article.
References
-
- Rahman MH, Biswas K, Hossain MA, Sack RB, Mekalanos JJ, Faruque SM. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 2008;27: 347–355. 10.1089/dna.2008.0737 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

