Intratumoral Heterogeneity of the Epigenome
- PMID: 27070699
- PMCID: PMC4852161
- DOI: 10.1016/j.ccell.2016.03.009
Intratumoral Heterogeneity of the Epigenome
Abstract
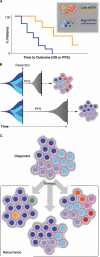
Investigation into intratumoral heterogeneity (ITH) of the epigenome is in a formative stage. The patterns of tumor evolution inferred from epigenetic ITH and genetic ITH are remarkably similar, suggesting widespread co-dependency of these disparate mechanisms. The biological and clinical relevance of epigenetic ITH are becoming more apparent. Rare tumor cells with unique and reversible epigenetic states may drive drug resistance, and the degree of epigenetic ITH at diagnosis may predict patient outcome. This perspective presents these current concepts and clinical implications of epigenetic ITH, and the experimental and computational techniques at the forefront of ITH exploration.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

References
-
- Aggerholm A, Guldberg P, Hokland M, Hokland P. Extensive intra- and interindividual heterogeneity of p15INK4B methylation in acute myeloid leukemia. Cancer Res. 1999;59:436–441. - PubMed
-
- Alpar D, Barber LJ, Gerlinger M. Chapter 24. Genetic Intratumor Heterogeneity. In: Gray SG, editor. Epigenetic Cancer Therapy. Elsevier Inc.; 2015. pp. 569–594.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
