Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs
- PMID: 27070701
- PMCID: PMC4889129
- DOI: 10.1016/j.ccell.2016.03.007
Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs
Abstract
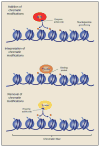
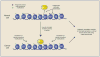
The epigenome is a key determinant of transcriptional output. Perturbations within the epigenome are thought to be a key feature of many, perhaps all cancers, and it is now clear that epigenetic changes are instrumental in cancer development. The inherent reversibility of these changes makes them attractive targets for therapeutic manipulation, and a number of small molecules targeting chromatin-based mechanisms are currently in clinical trials. In this perspective we discuss how understanding the cancer epigenome is providing insights into disease pathogenesis and informing drug development. We also highlight additional opportunities to further unlock the therapeutic potential within the cancer epigenome.
Keywords: Cancer; chromatin; epigenome; histone modification; targeted therapy.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, Nakitandwe J, Holmfeldt L, Parker M, Easton J, Huether R, Kriwacki R, Rusch M, Wu G, Li Y, Mulder H, Raimondi S, Pounds S, Kang G, Shi L, Becksfort J, Gupta P, Payne-Turner D, Vadodaria B, Boggs K, Yergeau D, Manne J, Song G, Edmonson M, Nagahawatte P, Wei L, Cheng C, Pei D, Sutton R, Venn NC, Chetcuti A, Rush A, Catchpoole D, Heldrup J, Fioretos T, Lu C, Ding L, Pui CH, Shurtleff S, Mullighan CG, Mardis ER, Wilson RK, Gruber Ta, Zhang J, Downing JR. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:1–147. doi: 10.1038/ng.3230. - DOI - PMC - PubMed
-
- Antonysamy S, Condon B, Druzina Z, Bonanno JB, Gheyi T, Zhang F, MacEwan I, Zhang A, Ashok S, Rodgers L, Russell M, Luz JG. Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-Ray crystallographic analysis of the EZH2-SET domain. PLoS One. 2013;8:1–15. doi: 10.1371/journal.pone.0084147. - DOI - PMC - PubMed
-
- Asangani Ia, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu YM, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. - DOI - PMC - PubMed
-
- Azuma K, Tsurutani J, Sakai K, Kaneda H, Fujisaka Y, Takeda M, Watatani M, Arao T, Satoh T, Okamoto I, Kurata T, Nishio K, Nakagawa K. Switching addictions between HER2 and FGFR2 in HER2-positive breast tumor cells: FGFR2 as a potential target for salvage after lapatinib failure. Biochem Biophys Res Commun. 2011;407:219–24. doi: 10.1016/j.bbrc.2011.03.002. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

