Pathogenetics of alveolar capillary dysplasia with misalignment of pulmonary veins
- PMID: 27071622
- PMCID: PMC5518754
- DOI: 10.1007/s00439-016-1655-9
Pathogenetics of alveolar capillary dysplasia with misalignment of pulmonary veins
Abstract
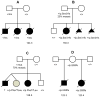
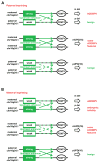
Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) is a lethal lung developmental disorder caused by heterozygous point mutations or genomic deletion copy-number variants (CNVs) of FOXF1 or its upstream enhancer involving fetal lung-expressed long noncoding RNA genes LINC01081 and LINC01082. Using custom-designed array comparative genomic hybridization, Sanger sequencing, whole exome sequencing (WES), and bioinformatic analyses, we studied 22 new unrelated families (20 postnatal and two prenatal) with clinically diagnosed ACDMPV. We describe novel deletion CNVs at the FOXF1 locus in 13 unrelated ACDMPV patients. Together with the previously reported cases, all 31 genomic deletions in 16q24.1, pathogenic for ACDMPV, for which parental origin was determined, arose de novo with 30 of them occurring on the maternally inherited chromosome 16, strongly implicating genomic imprinting of the FOXF1 locus in human lungs. Surprisingly, we have also identified four ACDMPV families with the pathogenic variants in the FOXF1 locus that arose on paternal chromosome 16. Interestingly, a combination of the severe cardiac defects, including hypoplastic left heart, and single umbilical artery were observed only in children with deletion CNVs involving FOXF1 and its upstream enhancer. Our data demonstrate that genomic imprinting at 16q24.1 plays an important role in variable ACDMPV manifestation likely through long-range regulation of FOXF1 expression, and may be also responsible for key phenotypic features of maternal uniparental disomy 16. Moreover, in one family, WES revealed a de novo missense variant in ESRP1, potentially implicating FGF signaling in the etiology of ACDMPV.
Conflict of interest statement
Figures



References
-
- Araujo E, Júnior, Palma-Dias R, Martins WP, Reidy K, da Silva Costa F. Congenital heart disease and adverse perinatal outcome in fetuses with confirmed isolated single functioning umbilical artery. J Obstet Gynaecol. 2015;35:85–87. - PubMed
-
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

