Predicting drug target interactions using meta-path-based semantic network analysis
- PMID: 27071755
- PMCID: PMC4830032
- DOI: 10.1186/s12859-016-1005-x
Predicting drug target interactions using meta-path-based semantic network analysis
Abstract
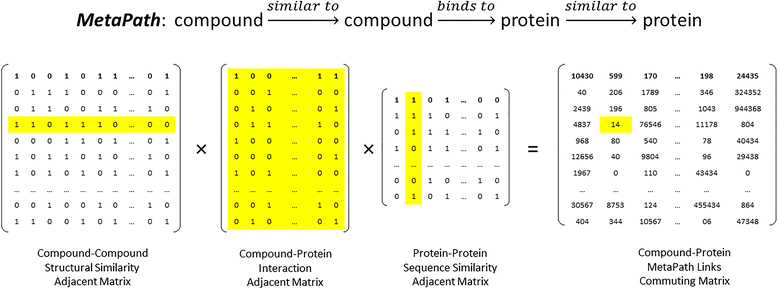
Background: In the context of drug discovery, drug target interactions (DTIs) can be predicted based on observed topological features of a semantic network across the chemical and biological space. In a semantic network, the types of the nodes and links are different. In order to take into account the heterogeneity of the semantic network, meta-path-based topological patterns were investigated for link prediction.
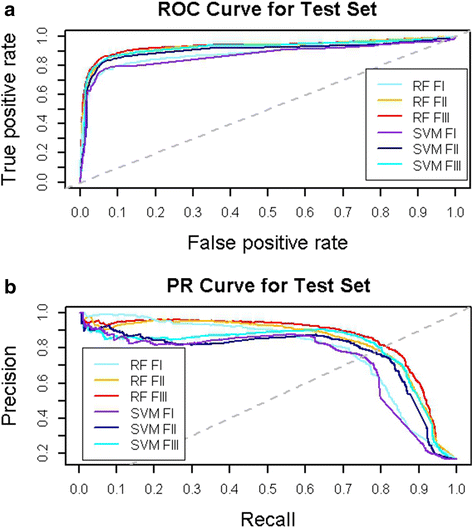
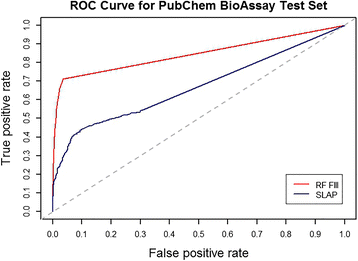
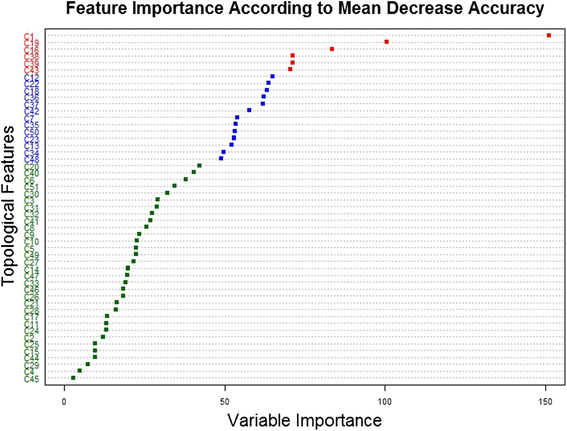
Results: Supervised machine learning models were constructed based on meta-path topological features of an enriched semantic network, which was derived from Chem2Bio2RDF, and was expanded by adding compound and protein similarity neighboring links obtained from the PubChem databases. The additional semantic links significantly improved the predictive performance of the supervised learning models. The binary classification model built upon the enriched feature space using the Random Forest algorithm significantly outperformed an existing semantic link prediction algorithm, Semantic Link Association Prediction (SLAP), to predict unknown links between compounds and protein targets in an evolving network. In addition to link prediction, Random Forest also has an intrinsic feature ranking algorithm, which can be used to select the important topological features that contribute to link prediction.
Conclusions: The proposed framework has been demonstrated as a powerful alternative to SLAP in order to predict DTIs using the semantic network that integrates chemical, pharmacological, genomic, biological, functional, and biomedical information into a unified framework. It offers the flexibility to enrich the feature space by using different normalization processes on the topological features, and it can perform model construction and feature selection at the same time.
Keywords: Link prediction; Machine learning; Meta-path topological feature; Random forest; Semantic network analysis.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

