Fast and sensitive taxonomic classification for metagenomics with Kaiju
- PMID: 27071849
- PMCID: PMC4833860
- DOI: 10.1038/ncomms11257
Fast and sensitive taxonomic classification for metagenomics with Kaiju
Abstract
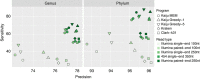
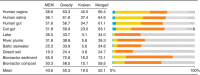
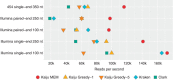
Metagenomics emerged as an important field of research not only in microbial ecology but also for human health and disease, and metagenomic studies are performed on increasingly larger scales. While recent taxonomic classification programs achieve high speed by comparing genomic k-mers, they often lack sensitivity for overcoming evolutionary divergence, so that large fractions of the metagenomic reads remain unclassified. Here we present the novel metagenome classifier Kaiju, which finds maximum (in-)exact matches on the protein-level using the Burrows-Wheeler transform. We show in a genome exclusion benchmark that Kaiju classifies reads with higher sensitivity and similar precision compared with current k-mer-based classifiers, especially in genera that are underrepresented in reference databases. We also demonstrate that Kaiju classifies up to 10 times more reads in real metagenomes. Kaiju can process millions of reads per minute and can run on a standard PC. Source code and web server are available at http://kaiju.binf.ku.dk.
Figures


References
-
- Riesenfeld C., Schloss P. & Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38, 525–552 (2004). - PubMed
-
- Shokralla S., Spall J., Gibson J. & Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 21, 1794–1805 (2012). - PubMed
-
- Kinross J., von Roon A., Holmes E., Darzi A. & Nicholson J. The human gut microbiome: implications for future health care. Curr. Gastroenterol. Rep. 10, 396–403 (2008). - PubMed
-
- Wade W. The oral microbiome in health and disease. Pharmacol. Res. 69, 137–143 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

