GPER signalling in both cancer-associated fibroblasts and breast cancer cells mediates a feedforward IL1β/IL1R1 response
- PMID: 27072893
- PMCID: PMC4829876
- DOI: 10.1038/srep24354
GPER signalling in both cancer-associated fibroblasts and breast cancer cells mediates a feedforward IL1β/IL1R1 response
Abstract
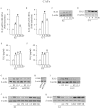
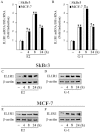
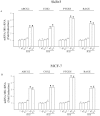
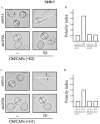
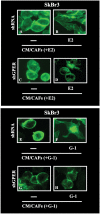
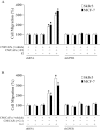
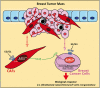
Cancer-associated fibroblasts (CAFs) contribute to the malignant aggressiveness through secreted factors like IL1β, which may drive pro-tumorigenic inflammatory phenotypes mainly acting via the cognate receptor named IL1R1. Here, we demonstrate that signalling mediated by the G protein estrogen receptor (GPER) triggers IL1β and IL1R1 expression in CAFs and breast cancer cells, respectively. Thereby, ligand-activation of GPER generates a feedforward loop coupling IL1β induction by CAFs to IL1R1 expression by cancer cells, promoting the up-regulation of IL1β/IL1R1 target genes such as PTGES, COX2, RAGE and ABCG2. This regulatory interaction between the two cell types induces migration and invasive features in breast cancer cells including fibroblastoid cytoarchitecture and F-actin reorganization. A better understanding of the mechanisms involved in the regulation of pro-inflammatory cytokines by GPER-integrated estrogen signals may be useful to target these stroma-cancer interactions.
Figures


Similar articles
-
GPER Mediates a Feedforward FGF2/FGFR1 Paracrine Activation Coupling CAFs to Cancer Cells toward Breast Tumor Progression.Cells. 2019 Mar 7;8(3):223. doi: 10.3390/cells8030223. Cells. 2019. PMID: 30866584 Free PMC article.
-
The IL1β-IL1R signaling is involved in the stimulatory effects triggered by hypoxia in breast cancer cells and cancer-associated fibroblasts (CAFs).J Exp Clin Cancer Res. 2020 Aug 10;39(1):153. doi: 10.1186/s13046-020-01667-y. J Exp Clin Cancer Res. 2020. PMID: 32778144 Free PMC article.
-
GPER activates Notch signaling in breast cancer cells and cancer-associated fibroblasts (CAFs).Int J Biochem Cell Biol. 2014 Jan;46:56-67. doi: 10.1016/j.biocel.2013.11.011. Epub 2013 Nov 22. Int J Biochem Cell Biol. 2014. PMID: 24275097
-
GPER is involved in the functional liaison between breast tumor cells and cancer-associated fibroblasts (CAFs).J Steroid Biochem Mol Biol. 2018 Feb;176:49-56. doi: 10.1016/j.jsbmb.2017.02.019. Epub 2017 Feb 27. J Steroid Biochem Mol Biol. 2018. PMID: 28249728 Review.
-
Crosstalk between Notch, HIF-1α and GPER in Breast Cancer EMT.Int J Mol Sci. 2018 Jul 10;19(7):2011. doi: 10.3390/ijms19072011. Int J Mol Sci. 2018. PMID: 29996493 Free PMC article. Review.
Cited by
-
Investigating the effect of Fusobacterium nucleatum on the aggressive behavior of cancer-associated fibroblasts in colorectal cancer.Discov Oncol. 2024 Jul 19;15(1):292. doi: 10.1007/s12672-024-01156-0. Discov Oncol. 2024. PMID: 39030445 Free PMC article.
-
GPER Mediates a Feedforward FGF2/FGFR1 Paracrine Activation Coupling CAFs to Cancer Cells toward Breast Tumor Progression.Cells. 2019 Mar 7;8(3):223. doi: 10.3390/cells8030223. Cells. 2019. PMID: 30866584 Free PMC article.
-
Low expression of G protein-coupled oestrogen receptor 1 (GPER) is associated with adverse survival of breast cancer patients.Oncotarget. 2018 May 25;9(40):25946-25956. doi: 10.18632/oncotarget.25408. eCollection 2018 May 25. Oncotarget. 2018. PMID: 29899833 Free PMC article.
-
The Expression of GPR30 in Iron-Overloaded Atypical Ovarian Epithelium and Ectopic Endometrium Is Closely Correlated with Transferrin Receptor and Pi3K.Biomed Res Int. 2022 Sep 12;2022:8338874. doi: 10.1155/2022/8338874. eCollection 2022. Biomed Res Int. 2022. PMID: 36132079 Free PMC article.
-
Carbon-Coated Magnetic Nanoparticle Dedicated to MRI/Photoacoustic Imaging of Tumor in Living Mice.Front Bioeng Biotechnol. 2021 Dec 2;9:800744. doi: 10.3389/fbioe.2021.800744. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34926438 Free PMC article.
References
-
- Erez N., Glanz S., Raz Y., Avivi C. & Barshack I. Cancer associated fibroblasts express pro-inflammatory factors in human breast and ovarian tumors. Biochem Biophys Res. Commun. 437, 397–402 (2013). - PubMed
-
- Han Y., Zhang Y., Jia T. & Sun Y. Molecular mechanism underlying the tumor-promoting functions of carcinoma-associated fibroblasts. Tumor Biol. 36, 1385–1394 (2015). - PubMed
-
- Kalluri R. & Zeisberg M. Fibroblasts in cancer. Nature Rev Cancer 6, 392–401 (2006). - PubMed
-
- Servais C. & Erez N. From sentinel cells to inflammatory culprits: cancer-associated fibroblasts in tumour-related inflammation. J. Pathol. 229, 198–207 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

