Role of Double-Strand Break End-Tethering during Gene Conversion in Saccharomyces cerevisiae
- PMID: 27074148
- PMCID: PMC4830573
- DOI: 10.1371/journal.pgen.1005976
Role of Double-Strand Break End-Tethering during Gene Conversion in Saccharomyces cerevisiae
Abstract
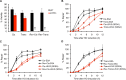
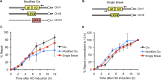
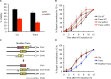
Correct repair of DNA double-strand breaks (DSBs) is critical for maintaining genome stability. Whereas gene conversion (GC)-mediated repair is mostly error-free, repair by break-induced replication (BIR) is associated with non-reciprocal translocations and loss of heterozygosity. We have previously shown that a Recombination Execution Checkpoint (REC) mediates this competition by preventing the BIR pathway from acting on DSBs that can be repaired by GC. Here, we asked if the REC can also determine whether the ends that are engaged in a GC-compatible configuration belong to the same break, since repair involving ends from different breaks will produce potentially deleterious translocations. We report that the kinetics of repair are markedly delayed when the two DSB ends that participate in GC belong to different DSBs (termed Trans) compared to the case when both DSB ends come from the same break (Cis). However, repair in Trans still occurs by GC rather than BIR, and the overall efficiency of repair is comparable. Hence, the REC is not sensitive to the "origin" of the DSB ends. When the homologous ends for GC are in Trans, the delay in repair appears to reflect their tethering to sequences on the other side of the DSB that themselves recombine with other genomic locations with which they share sequence homology. These data support previous observations that the two ends of a DSB are usually tethered to each other and that this tethering facilitates both ends encountering the same donor sequence. We also found that the presence of homeologous/repetitive sequences in the vicinity of a DSB can distract the DSB end from finding its bona fide homologous donor, and that inhibition of GC by such homeologous sequences is markedly increased upon deleting Sgs1 but not Msh6.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271. - PubMed
-
- McEachern MJ, Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75: 111–135. - PubMed
-
- Haber JE (2013) Genome Stability: DNA Repair and Recombination: Garland Science. 396 p.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

