Mutation-specific effects on thin filament length in thin filament myopathy
- PMID: 27074222
- PMCID: PMC4911820
- DOI: 10.1002/ana.24654
Mutation-specific effects on thin filament length in thin filament myopathy
Abstract
Objective: Thin filament myopathies are among the most common nondystrophic congenital muscular disorders, and are caused by mutations in genes encoding proteins that are associated with the skeletal muscle thin filament. Mechanisms underlying muscle weakness are poorly understood, but might involve the length of the thin filament, an important determinant of force generation.
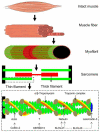
Methods: We investigated the sarcomere length-dependence of force, a functional assay that provides insights into the contractile strength of muscle fibers as well as the length of the thin filaments, in muscle fibers from 51 patients with thin filament myopathy caused by mutations in NEB, ACTA1, TPM2, TPM3, TNNT1, KBTBD13, KLHL40, and KLHL41.
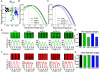
Results: Lower force generation was observed in muscle fibers from patients of all genotypes. In a subset of patients who harbor mutations in NEB and ACTA1, the lower force was associated with downward shifted force-sarcomere length relations, indicative of shorter thin filaments. Confocal microscopy confirmed shorter thin filaments in muscle fibers of these patients. A conditional Neb knockout mouse model, which recapitulates thin filament myopathy, revealed a compensatory mechanism; the lower force generation that was associated with shorter thin filaments was compensated for by increasing the number of sarcomeres in series. This allowed muscle fibers to operate at a shorter sarcomere length and maintain optimal thin-thick filament overlap.
Interpretation: These findings might provide a novel direction for the development of therapeutic strategies for thin filament myopathy patients with shortened thin filament lengths. Ann Neurol 2016;79:959-969.
© 2016 American Neurological Association.
Figures


References
-
- Sanoudou D, Beggs AH. Clinical and genetic heterogeneity in nemaline myopathy—a disease of skeletal muscle thin filaments. Trends Mol Med. 2001;7:362–368. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

