On the Quantification of Cellular Velocity Fields
- PMID: 27074673
- PMCID: PMC4833777
- DOI: 10.1016/j.bpj.2016.02.032
On the Quantification of Cellular Velocity Fields
Abstract
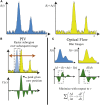
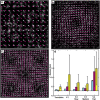
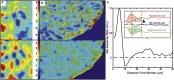
The application of flow visualization in biological systems is becoming increasingly common in studies ranging from intracellular transport to the movements of whole organisms. In cell biology, the standard method for measuring cell-scale flows and/or displacements has been particle image velocimetry (PIV); however, alternative methods exist, such as optical flow constraint. Here we review PIV and optical flow, focusing on the accuracy and efficiency of these methods in the context of cellular biophysics. Although optical flow is not as common, a relatively simple implementation of this method can outperform PIV and is easily augmented to extract additional biophysical/chemical information such as local vorticity or net polymerization rates from speckle microscopy.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Danuser G., Waterman-Storer C.M. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 2006;35:361–387. - PubMed
-
- Bise R., Kanade T., Yin Z., Huh S.I. Automatic cell tracking applied to analysis of cell migration in wound healing assay. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2011;2011:6174–6179. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

