Structure, function, and genetics of lipoprotein (a)
- PMID: 27074913
- PMCID: PMC4959873
- DOI: 10.1194/jlr.R067314
Structure, function, and genetics of lipoprotein (a)
Abstract
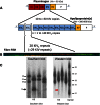
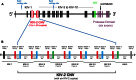
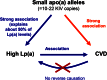
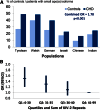
Lipoprotein (a) [Lp(a)] has attracted the interest of researchers and physicians due to its intriguing properties, including an intragenic multiallelic copy number variation in the LPA gene and the strong association with coronary heart disease (CHD). This review summarizes present knowledge of the structure, function, and genetics of Lp(a) with emphasis on the molecular and population genetics of the Lp(a)/LPA trait, as well as aspects of genetic epidemiology. It highlights the role of genetics in establishing Lp(a) as a risk factor for CHD, but also discusses uncertainties, controversies, and lack of knowledge on several aspects of the genetic Lp(a) trait, not least its function.
Keywords: Mendelian randomization; cardiovascular risk factor; copy number variation; lipoprotein metabolism.
Copyright © 2016 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Berg K. 1963. A new serum type system in man–the Lp system. Acta Pathol. Microbiol. Scand. 59: 369–382. - PubMed
-
- Kronenberg F., and Utermann G.. 2013. Lipoprotein(a): resurrected by genetics. J. Intern. Med. 273: 6–30. - PubMed
-
- Ehnholm C., Garoff H., Renkonen O., and Simons K.. 1972. Protein and carbohydrate composition of Lp(a)lipoprotein from human plasma. Biochemistry. 11: 3229–3232. - PubMed
-
- Gaubatz J. W., Heideman C., Gotto A. M. Jr., Morrisett J. D., and Dahlen G. H.. 1983. Human plasma lipoprotein(a): structural properties. J. Biol. Chem. 258: 4582–4589. - PubMed
-
- Utermann G., and Weber W.. 1983. Protein composition of Lp(a) lipoprotein from human plasma. FEBS Lett. 154: 357–361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

