Lipopolysaccharide modification in Gram-negative bacteria during chronic infection
- PMID: 27075488
- PMCID: PMC4931227
- DOI: 10.1093/femsre/fuw007
Lipopolysaccharide modification in Gram-negative bacteria during chronic infection
Abstract
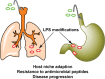
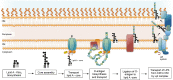
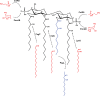
The Gram-negative bacterial lipopolysaccharide (LPS) is a major component of the outer membrane that plays a key role in host-pathogen interactions with the innate immune system. During infection, bacteria are exposed to a host environment that is typically dominated by inflammatory cells and soluble factors, including antibiotics, which provide cues about regulation of gene expression. Bacterial adaptive changes including modulation of LPS synthesis and structure are a conserved theme in infections, irrespective of the type or bacteria or the site of infection. In general, these changes result in immune system evasion, persisting inflammation and increased antimicrobial resistance. Here, we review the modifications of LPS structure and biosynthetic pathways that occur upon adaptation of model opportunistic pathogens (Pseudomonas aeruginosa, Burkholderia cepacia complex bacteria, Helicobacter pylori and Salmonella enterica) to chronic infection in respiratory and gastrointestinal sites. We also discuss the molecular mechanisms of these variations and their role in the host-pathogen interaction.
Keywords: Burkholderia cenocepacia; Helicobacter pylori; O antigen; Pseudomonas aeruginosa; adaptive mutation; cystic fibrosis; gastric ulcer; lipid A.
© FEMS 2016. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Aaron SD, Vandemheen KL, Ramotar K, et al. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA. 2010;304:2145–53. - PubMed
-
- Aspinall GO, Monteiro MA, Pang H, et al. Lipopolysaccharide of the Helicobacter pylori type strain NCTC 11637 (ATCC 43504): structure of the O antigen chain and core oligosaccharide regions. Biochemistry. 1996;35:2489–97. - PubMed
-
- Azmuda N, Rahman MZ, Sultana M, et al. Evidence of interspecies O antigen gene cluster transfer between Shigella boydii 15 and Escherichia fergusonii. APMIS. 2012;120:959–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

