Alternative promoters of Peg3 with maternal specificity
- PMID: 27075691
- PMCID: PMC4830991
- DOI: 10.1038/srep24438
Alternative promoters of Peg3 with maternal specificity
Abstract
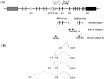
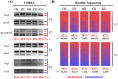
Peg3 (paternally expressed gene 3) is an imprinted gene localized within an evolutionarily conserved 500-kb domain in human chromosome 19q13.4 and mouse proximal chromosome 7. In the current study, we have identified three alternative promoters for mouse Peg3 and one alternative promoter for human PEG3. These alternative promoters are localized within the 200-kb upstream region of human and mouse PEG3, which is well conserved and thus predicted to harbor several cis-regulatory elements for the PEG3 domain. In the mouse, two of these alternative promoters drive maternal-specific expression of Peg3 specifically in the hypothalamus of the adult brain, while the remaining third promoter drives bi-allelic expression of Peg3 with a paternal bias only in the neonatal-stage brain. In human, an alternative transcript is also detected at relatively very low levels in adult brain and placenta. Overall, the identification of alternative promoters in both mouse and human models suggests that these alternative promoters may be functionally selected features for the PEG3 imprinted domain during mammalian evolution.
Figures




Similar articles
-
Sex and Tissue Specificity of Peg3 Promoters.PLoS One. 2016 Oct 6;11(10):e0164158. doi: 10.1371/journal.pone.0164158. eCollection 2016. PLoS One. 2016. PMID: 27711129 Free PMC article.
-
Identification of an evolutionarily conserved cis-regulatory element controlling the Peg3 imprinted domain.PLoS One. 2013 Sep 10;8(9):e75417. doi: 10.1371/journal.pone.0075417. eCollection 2013. PLoS One. 2013. PMID: 24040411 Free PMC article.
-
The human homolog of a mouse-imprinted gene, Peg3, maps to a zinc finger gene-rich region of human chromosome 19q13.4.Genome Res. 1997 May;7(5):532-40. doi: 10.1101/gr.7.5.532. Genome Res. 1997. PMID: 9149948 Free PMC article.
-
Regulation and function of the peg3 imprinted domain.Genomics Inform. 2014 Sep;12(3):105-13. doi: 10.5808/GI.2014.12.3.105. Epub 2014 Sep 30. Genomics Inform. 2014. PMID: 25317109 Free PMC article. Review.
-
Organization of the human aromatase p450 (CYP19) gene.Semin Reprod Med. 2004 Feb;22(1):5-9. doi: 10.1055/s-2004-823022. Semin Reprod Med. 2004. PMID: 15083376 Review.
Cited by
-
Transcription factor expression defines subclasses of developing projection neurons highly similar to single-cell RNA-seq subtypes.Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):25074-25084. doi: 10.1073/pnas.2008013117. Epub 2020 Sep 18. Proc Natl Acad Sci U S A. 2020. PMID: 32948690 Free PMC article.
-
Long noncoding RNA ZNF667-AS1 reduces tumor invasion and metastasis in cervical cancer by counteracting microRNA-93-3p-dependent PEG3 downregulation.Mol Oncol. 2019 Nov;13(11):2375-2392. doi: 10.1002/1878-0261.12565. Epub 2019 Oct 17. Mol Oncol. 2019. Retraction in: Mol Oncol. 2025 Jan;19(1):263. doi: 10.1002/1878-0261.13777. PMID: 31420931 Free PMC article. Retracted.
-
The imprinted gene Pw1/Peg3 regulates skeletal muscle growth, satellite cell metabolic state, and self-renewal.Sci Rep. 2018 Oct 2;8(1):14649. doi: 10.1038/s41598-018-32941-x. Sci Rep. 2018. PMID: 30279563 Free PMC article.
-
Circular RNA identified from Peg3 and Igf2r.PLoS One. 2018 Sep 14;13(9):e0203850. doi: 10.1371/journal.pone.0203850. eCollection 2018. PLoS One. 2018. PMID: 30216384 Free PMC article.
-
Parental and sexual conflicts over the Peg3 imprinted domain.Sci Rep. 2016 Nov 30;6:38136. doi: 10.1038/srep38136. Sci Rep. 2016. PMID: 27901122 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

