Reliable handling of highly A/T-rich genomic DNA for efficient generation of knockin strains of Dictyostelium discoideum
- PMID: 27075750
- PMCID: PMC4831088
- DOI: 10.1186/s12896-016-0267-8
Reliable handling of highly A/T-rich genomic DNA for efficient generation of knockin strains of Dictyostelium discoideum
Abstract
Background: Social amoeba, Dictyostelium discoideum, is a well-established model organism for studying cellular physiology and developmental pattern formation. Its haploid genome facilitates functional analysis of genes by a single round of mutagenesis including targeted disruption. Although the efficient generation of knockout strains based on an intrinsically high homologous recombination rate has been demonstrated, successful reports for knockin strains have been limited. As social amoeba has an exceptionally high adenine and thymine (A/T)-content, conventional plasmid-based vector construction has been constrained due to deleterious deletion in E. coli.
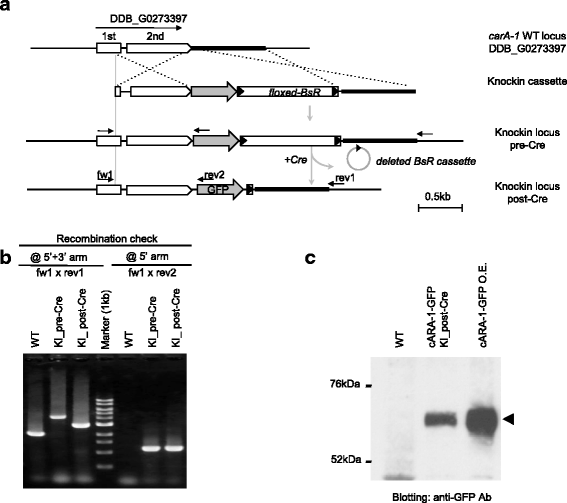
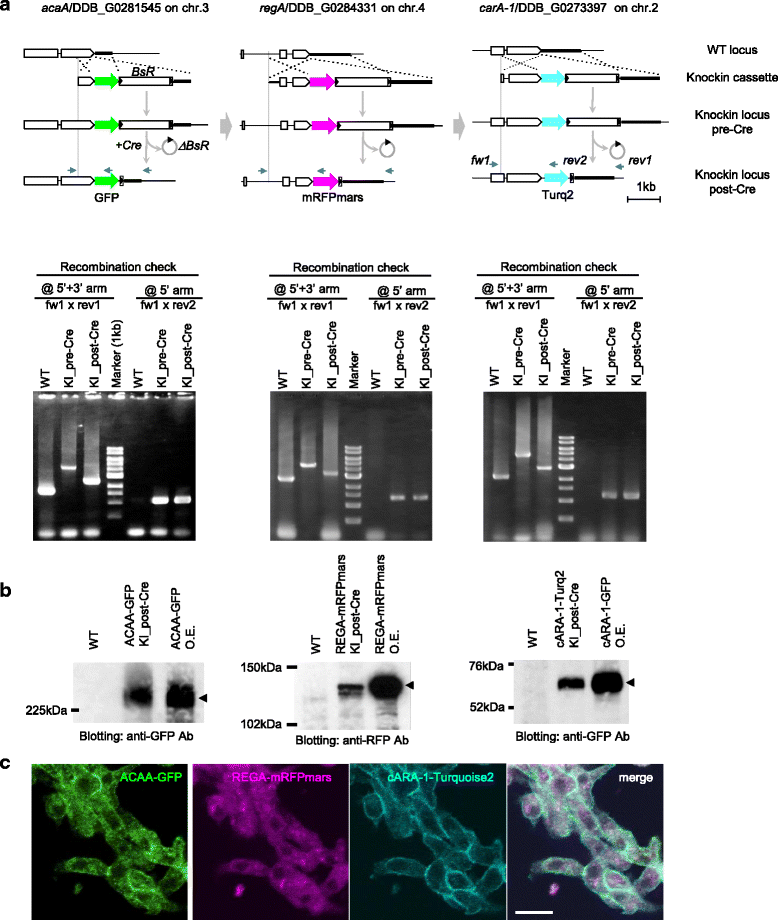
Results: We describe here a simple and efficient strategy to construct GFP-knockin cassettes by using a linear DNA cloning vector derived from N15 bacteriophage. This allows reliable handling of DNA fragments whose A/T-content may be as high as 85 %, and which cannot be cloned into a circular plasmid. By optimizing the length of recombination arms, we successfully generate GFP-knockin strains for five genes involved in cAMP signalling, including a triple-colour knockin strain.
Conclusions: This robust strategy would be useful in handling DNA fragments with biased A/T-contents such as the genome of lower organisms and the promoter/terminator regions of higher organisms.
Keywords: A/T-rich genome; Dictyostelium discoideum; Knockin; Linear DNA cloning.
Figures

Similar articles
-
The application of the Cre-loxP system for generating multiple knock-out and knock-in targeted loci.Methods Mol Biol. 2013;983:249-67. doi: 10.1007/978-1-62703-302-2_13. Methods Mol Biol. 2013. PMID: 23494311
-
Efficient generation of gene knockout plasmids for Dictyostelium discoideum using one-step cloning.Genomics. 2011 May;97(5):321-5. doi: 10.1016/j.ygeno.2011.02.001. Epub 2011 Feb 19. Genomics. 2011. PMID: 21316445
-
Efficient circularization in Escherichia coli of linear plasmid multimers from Dictyostelium discoideum genomic DNA.Plasmid. 1996 Sep;36(2):86-94. doi: 10.1006/plas.1996.0036. Plasmid. 1996. PMID: 8954880
-
Dictyostelium genomics: how it developed and what we have learned from it.Pharmazie. 2013 Jul;68(7):474-7. Pharmazie. 2013. PMID: 23923625 Review.
-
Transfer RNA gene-targeted integration: an adaptation of retrotransposable elements to survive in the compact Dictyostelium discoideum genome.Cytogenet Genome Res. 2005;110(1-4):288-98. doi: 10.1159/000084961. Cytogenet Genome Res. 2005. PMID: 16093681 Review.
Cited by
-
Knock-in and precise nucleotide substitution using near-PAMless engineered Cas9 variants in Dictyostelium discoideum.Sci Rep. 2021 May 27;11(1):11163. doi: 10.1038/s41598-021-89546-0. Sci Rep. 2021. PMID: 34045481 Free PMC article.
-
Multi-color fluorescence live-cell imaging in Dictyostelium discoideum.Cell Struct Funct. 2024 Dec 27;49(2):135-153. doi: 10.1247/csf.24065. Epub 2024 Dec 4. Cell Struct Funct. 2024. PMID: 39631875 Free PMC article.
-
When Dicty Met Myco, a (Not So) Romantic Story about One Amoeba and Its Intracellular Pathogen.Front Cell Infect Microbiol. 2018 Jan 9;7:529. doi: 10.3389/fcimb.2017.00529. eCollection 2017. Front Cell Infect Microbiol. 2018. PMID: 29376033 Free PMC article. Review.
-
A GoldenBraid cloning system for synthetic biology in social amoebae.Nucleic Acids Res. 2020 May 7;48(8):4139-4146. doi: 10.1093/nar/gkaa185. Nucleic Acids Res. 2020. PMID: 32232356 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

