Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation
- PMID: 27076641
- PMCID: PMC4886785
- DOI: 10.1128/JVI.00394-16
Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation
Abstract
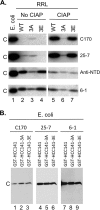
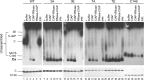
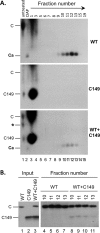
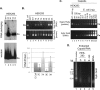
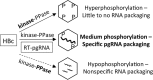
Multiple subunits of the hepatitis B virus (HBV) core protein (HBc) assemble into an icosahedral capsid that packages the viral pregenomic RNA (pgRNA). The N-terminal domain (NTD) of HBc is sufficient for capsid assembly, in the absence of pgRNA or any other viral or host factors, under conditions of high HBc and/or salt concentrations. The C-terminal domain (CTD) is deemed dispensable for capsid assembly although it is essential for pgRNA packaging. We report here that HBc expressed in a mammalian cell lysate, rabbit reticulocyte lysate (RRL), was able to assemble into capsids when (low-nanomolar) HBc concentrations mimicked those achieved under conditions of viral replication in vivo and were far below those used previously for capsid assembly in vitro Furthermore, at physiologically low HBc concentrations in RRL, the NTD was insufficient for capsid assembly and the CTD was also required. The CTD likely facilitated assembly under these conditions via RNA binding and protein-protein interactions. Moreover, the CTD underwent phosphorylation and dephosphorylation events in RRL similar to those seen in vivo which regulated capsid assembly. Importantly, the NTD alone also failed to accumulate in mammalian cells, likely resulting from its failure to assemble efficiently. Coexpression of the full-length HBc rescued NTD assembly in RRL as well as NTD expression and assembly in mammalian cells, resulting in the formation of mosaic capsids containing both full-length HBc and the NTD. These results have important implications for HBV assembly during replication and provide a facile cell-free system to study capsid assembly under physiologically relevant conditions, including its modulation by host factors.
Importance: Hepatitis B virus (HBV) is an important global human pathogen and the main cause of liver cancer worldwide. An essential component of HBV is the spherical capsid composed of multiple copies of a single protein, the core protein (HBc). We have developed a mammalian cell-free system in which HBc is expressed at physiological (low) concentrations and assembles into capsids under near-physiological conditions. In this cell-free system, as in mammalian cells, capsid assembly depends on the C-terminal domain (CTD) of HBc, in contrast to other assembly systems in which HBc assembles into capsids independently of the CTD under conditions of nonphysiological protein and salt concentrations. Furthermore, the phosphorylation state of the CTD regulates capsid assembly and RNA encapsidation in the cell-free system in a manner similar to that seen in mammalian cells. This system will facilitate detailed studies on capsid assembly and RNA encapsidation under physiological conditions and identification of antiviral agents that target HBc.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Seeger C, Zoulim F, Mason WS. 2013. Hepadnaviruses, p 2185–2221. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott, Williams & Wilkins, Philadelphia, PA.
-
- Hu J. 2016. Hepatitis B virus virology and replication, p 1–34. In Liaw Y-F, Zoulim F (ed), Hepatitis B virus in human diseases. Humana Press, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

