Proximity assays for sensitive quantification of proteins
- PMID: 27077033
- PMCID: PMC4822221
- DOI: 10.1016/j.bdq.2015.04.002
Proximity assays for sensitive quantification of proteins
Abstract
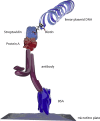
Proximity assays are immunohistochemical tools that utilise two or more DNA-tagged aptamers or antibodies binding in close proximity to the same protein or protein complex. Amplification by PCR or isothermal methods and hybridisation of a labelled probe to its DNA target generates a signal that enables sensitive and robust detection of proteins, protein modifications or protein-protein interactions. Assays can be carried out in homogeneous or solid phase formats and in situ assays can visualise single protein molecules or complexes with high spatial accuracy. These properties highlight the potential of proximity assays in research, diagnostic, pharmacological and many other applications that require sensitive, specific and accurate assessments of protein expression.
Keywords: Immuno-PCR; Immunoassays; In situ assays; Proximity extension assay; Proximity ligation assay.
Figures
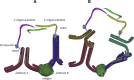


References
-
- Bustin S.A. IUL Press; La Jolla, CA: 2004. A–Z of quantitative PCR.
-
- Higuchi R., Fockler C., Dollinger G., Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 1993;11:1026–1030. - PubMed
-
- Gibson U.E., Heid C.A., Williams P.M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. - PubMed
-
- Heid C.A., Stevens J., Livak K.J., Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. - PubMed
-
- Engvall E., Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
