Differential amplicons (ΔAmp)-a new molecular method to assess RNA integrity
- PMID: 27077042
- PMCID: PMC4822209
- DOI: 10.1016/j.bdq.2015.09.002
Differential amplicons (ΔAmp)-a new molecular method to assess RNA integrity
Abstract
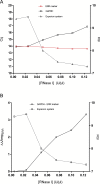
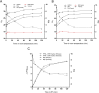
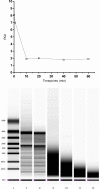
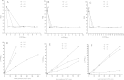
Integrity of the mRNA in clinical samples has major impact on the quality of measured expression levels. This is independent of the measurement technique being next generation sequencing (NGS), Quantitative real-time PCR (qPCR) or microarray profiling. If mRNA is highly degraded or damaged, measured data will be very unreliable and the whole study is likely a waste of time and money. It is therefore common strategy to test the quality of RNA in samples before conducting large and costly studies. Most methods today to assess the quality of RNA are ignorant to the nature of the RNA and, therefore, reflect the integrity of ribosomal RNA, which is the dominant species, rather than of mRNAs, microRNAs and long non-coding RNAs, which usually are the species of interest. Here, we present a novel molecular approach to assess the quality of the targeted RNA species by measuring the differential amplification (ΔAmp) of an Endogenous RNase Resistant (ERR) marker relative to a reference gene, optionally combined with the measurement of two amplicons of different lengths. The combination reveals any mRNA degradation caused by ribonucleases as well as physical, chemical or UV damage. ΔAmp has superior sensitivity to common microfluidic electrophoretic methods, senses the integrity of the actual targeted RNA species, and allows for a smoother and more cost efficient workflow.
Keywords: DAmp; Differential amplicons; Endogenous RNase resistant marker; RNA integrity; RNA quality; ΔAmp.
Figures
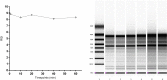
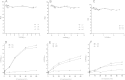

References
-
- Tichopad A., Kitchen R., Riedmaier I., Becker C., Stahlberg A., Kubista M. Design and optimization of reverse-transcription quantitative pcr experiments. Clin. Chem. 2009;55:101816–101823. - PubMed
-
- Pazzagli M., Malentacchi F., Simi L., Orlando C., Wyrich R., Günther K. SPIDIA-RNA: first external quality assessment for the pre-analytical phase of blood samples used for RNA based analyses. Methods. 2013;59:20–31. - PubMed
-
- Li Y., Breaker R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121(23):5364–5372.
-
- Larson D.E., Zahradka P., Sells B.H. Control points in eukaryotic ribosome biogenesis. Biochem. Cell Biol. 1991;69:5–22. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
