Incidentalome from Genomic Sequencing: A Barrier to Personalized Medicine?
- PMID: 27077130
- PMCID: PMC4816806
- DOI: 10.1016/j.ebiom.2016.01.030
Incidentalome from Genomic Sequencing: A Barrier to Personalized Medicine?
Abstract
Background: In Western cohorts, the prevalence of incidental findings (IFs) or incidentalome, referring to variants in genes that are unrelated to the patient's primary condition, is between 0.86% and 8.8%. However, data on prevalence and type of IFs in Asian population is lacking.
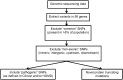
Methods: In 2 cohorts of individuals with genomic sequencing performed in Singapore (total n = 377), we extracted and annotated variants in the 56 ACMG-recommended genes and filtered these variants based on the level of pathogenicity. We then analyzed the precise distribution of IFs, class of genes, related medical conditions, and potential clinical impact.
Results: We found a total of 41,607 variants in the 56 genes in our cohort of 377 individuals. After filtering for rare and coding variants, we identified 14 potential variants. After reviewing primary literature, only 4 out of the 14 variants were classified to be pathogenic, while an additional two variants were classified as likely pathogenic. Overall, the cumulative prevalence of IFs (pathogenic and likely pathogenic variants) in our cohort was 1.6%.
Conclusion: The cumulative prevalence of IFs through genomic sequencing is low and the incidentalome may not be a significant barrier to implementation of genomics for personalized medicine.
Keywords: Genomic sequencing; Incidental findings; Personalized medicine.
References
-
- Ayuso C., Millan J.M., Dal-Re R. Management and return of incidental genomic findings in clinical trials. Pharmacogenomics J. 2015;15(1):1–5. - PubMed
-
- Ballinger M.L., Mitchell G., Thomas D.M. Surveillance recommendations for patients with germline TP53 mutations. Curr. Opin. Oncol. 2015;27(4):332–337. - PubMed
-
- Berg J.S., Khoury M.J., Evans J.P. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet. Med. 2011;13(6):499–504. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources