Identification of Genome-Wide Variants and Discovery of Variants Associated with Brassica rapa Clubroot Resistance Gene Rcr1 through Bulked Segregant RNA Sequencing
- PMID: 27078023
- PMCID: PMC4831815
- DOI: 10.1371/journal.pone.0153218
Identification of Genome-Wide Variants and Discovery of Variants Associated with Brassica rapa Clubroot Resistance Gene Rcr1 through Bulked Segregant RNA Sequencing
Abstract
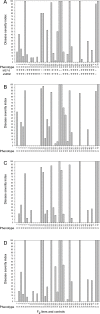
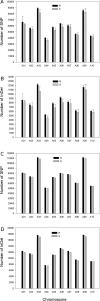
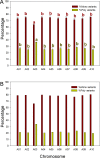
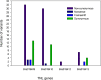
Clubroot, caused by Plasmodiophora brassicae, is an important disease on Brassica species worldwide. A clubroot resistance gene, Rcr1, with efficacy against pathotype 3 of P. brassicae, was previously mapped to chromosome A03 of B. rapa in pak choy cultivar "Flower Nabana". In the current study, resistance to pathotypes 2, 5 and 6 was shown to be associated with Rcr1 region on chromosome A03. Bulked segregant RNA sequencing was performed and short read sequences were assembled into 10 chromosomes of the B. rapa reference genome v1.5. For the resistant (R) bulks, a total of 351.8 million (M) sequences, 30,836.5 million bases (Mb) in length, produced 120-fold coverage of the reference genome. For the susceptible (S) bulks, 322.9 M sequences, 28,216.6 Mb in length, produced 109-fold coverage. In total, 776.2 K single nucleotide polymorphisms (SNPs) and 122.2 K insertion / deletion (InDels) in R bulks and 762.8 K SNPs and 118.7 K InDels in S bulks were identified; each chromosome had about 87% SNPs and 13% InDels, with 78% monomorphic and 22% polymorphic variants between the R and S bulks. Polymorphic variants on each chromosome were usually below 23%, but made up 34% of the variants on chromosome A03. There were 35 genes annotated in the Rcr1 target region and variants were identified in 21 genes. The numbers of poly variants differed significantly among the genes. Four out of them encode Toll-Interleukin-1 receptor / nucleotide-binding site / leucine-rich-repeat proteins; Bra019409 and Bra019410 harbored the higher numbers of polymorphic variants, which indicates that they are more likely candidates of Rcr1. Fourteen SNP markers in the target region were genotyped using the Kompetitive Allele Specific PCR method and were confirmed to associate with Rcr1. Selected SNP markers were analyzed with 26 recombinants obtained from a segregating population consisting of 1587 plants, indicating that they were completely linked to Rcr1. Nine SNP markers were used for marker-assisted introgression of Rcr1 into B. napus canola from B. rapa, with 100% accuracy in this study.
Conflict of interest statement
Figures


References
-
- Dixon GR. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J Plant Growth Regul 2009; 28; 194–202. 10.1007/s00344-009-9090-y - DOI
-
- Howard RJ, Strelkov SE, Hardinget MW.Clubroot of cruciferous crops–new perspectives on an old disease. Can. J. Plant Pathol 2010; 32: 43–57. 10.1080/07060661003621761 - DOI
-
- Kageyama K, Asano T. Life cycle of Plasmodiophora brassicae. J Plant Growth Regul 2009; 28: 203–11. 10.1007/s00344-009-9101-z - DOI
-
- Diederichsen E, Frauen M, Linders EGA, Hatakeyama EK, Hirai EM. Status and perspectives of clubroot resistance breeding in crucifer crops. J Plant Growth Regul 2009; 28:265–281. 10.1007/s00344-009-9100-0 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

