Evolution of Epigenetic Regulation in Vertebrate Genomes
- PMID: 27080453
- PMCID: PMC4842087
- DOI: 10.1016/j.tig.2016.03.001
Evolution of Epigenetic Regulation in Vertebrate Genomes
Abstract
Empirical models of sequence evolution have spurred progress in the field of evolutionary genetics for decades. We are now realizing the importance and complexity of the eukaryotic epigenome. While epigenome analysis has been applied to genomes from single-cell eukaryotes to human, comparative analyses are still relatively few and computational algorithms to quantify epigenome evolution remain scarce. Accordingly, a quantitative model of epigenome evolution remains to be established. We review here the comparative epigenomics literature and synthesize its overarching themes. We also suggest one mechanism, transcription factor binding site (TFBS) turnover, which relates sequence evolution to epigenetic conservation or divergence. Lastly, we propose a framework for how the field can move forward to build a coherent quantitative model of epigenome evolution.
Keywords: epigenome evolution; gene regulation; vertebrate genomics.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures



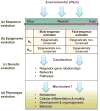
References
-
- Lukas J, et al. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. - PubMed
-
- Okamoto I, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. - PubMed
-
- Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Current Opinion in Cell Biology. 2009;21:359–366. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

