Epigenetic regulation of bone cells
- PMID: 27082893
- PMCID: PMC5498111
- DOI: 10.1080/03008207.2016.1177037
Epigenetic regulation of bone cells
Abstract
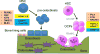
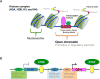
Bone is a major organ in the skeletal system that supports and protects muscles and other organs, facilitates movement and hematopoiesis, and forms a reservoir of minerals including calcium. The cells in the bone, such as osteoblasts, osteoclasts, and osteocytes, orchestrate sequential and balanced regulatory mechanisms to maintain bone and are capable of differentiating in bones. Bone development and remodeling require a precise regulation of gene expressions in bone cells, a process governed by epigenetic mechanisms such as histone modification, DNA methylation, and chromatin structure. Importantly, lineage-specific transcription factors can determine the epigenetic regulation of bone cells. Emerging data suggest that perturbation of epigenetic programs can affect the function and activity of bone cells and contributes to pathogenesis of bone diseases, including osteoporosis. Thus, understanding epigenetic regulations in bone cells would be important for early diagnosis and future therapeutic approaches.
Keywords: DNA methylation; epigenetic regulation; histone modification; osteoblasts; osteoclasts.
Conflict of interest statement
The author reports no conflicts of interest. The author alone is responsible for the content and writing of the article.
Figures
Similar articles
-
Epigenetic Regulation of Skeletal Tissue Integrity and Osteoporosis Development.Int J Mol Sci. 2020 Jul 12;21(14):4923. doi: 10.3390/ijms21144923. Int J Mol Sci. 2020. PMID: 32664681 Free PMC article. Review.
-
[Regulation of bone homeostasis by bone cells].Clin Calcium. 2013 Feb;23(2):218-28. Clin Calcium. 2013. PMID: 23354089 Review. Japanese.
-
Epigenetic mechanisms in bone.Clin Chem Lab Med. 2014 May;52(5):589-608. doi: 10.1515/cclm-2013-0770. Clin Chem Lab Med. 2014. PMID: 24353145 Review.
-
The emerging role of microRNAs in bone remodeling and its therapeutic implications for osteoporosis.Biosci Rep. 2018 Jun 21;38(3):BSR20180453. doi: 10.1042/BSR20180453. Print 2018 Jun 29. Biosci Rep. 2018. PMID: 29848766 Free PMC article. Review.
-
Quantitative analysis of the bone destroying activity of osteocytes and osteoclasts in experimental disuse osteoporosis.Ital J Orthop Traumatol. 1979 Aug;5(2):225-40. Ital J Orthop Traumatol. 1979. PMID: 548516
Cited by
-
Epigenetic Regulation of ZNF687 by miR-142a-3p and DNA Methylation During Osteoblast Differentiation and Mice Bone Development and Aging.Int J Mol Sci. 2025 Feb 27;26(5):2069. doi: 10.3390/ijms26052069. Int J Mol Sci. 2025. PMID: 40076693 Free PMC article.
-
Epigenetic roles of chromatin remodeling complexes in bone biology and the pathogenesis of bone‑related disease (Review).Int J Mol Med. 2025 Aug;56(2):115. doi: 10.3892/ijmm.2025.5556. Epub 2025 May 30. Int J Mol Med. 2025. PMID: 40444490 Free PMC article. Review.
-
Nuclear mechanosensing controls MSC osteogenic potential through HDAC epigenetic remodeling.Proc Natl Acad Sci U S A. 2020 Sep 1;117(35):21258-21266. doi: 10.1073/pnas.2006765117. Epub 2020 Aug 17. Proc Natl Acad Sci U S A. 2020. PMID: 32817542 Free PMC article.
-
Epigenetic Control of Osteogenic Lineage Commitment.Front Cell Dev Biol. 2021 Jan 8;8:611197. doi: 10.3389/fcell.2020.611197. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33490076 Free PMC article. Review.
-
The KDM4B-CCAR1-MED1 axis is a critical regulator of osteoclast differentiation and bone homeostasis.Bone Res. 2021 May 25;9(1):27. doi: 10.1038/s41413-021-00145-1. Bone Res. 2021. PMID: 34031372 Free PMC article.
References
-
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nature reviews Molecular cell biology. 2009;10:192–206. - PubMed
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33(Suppl):245–54. - PubMed
-
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. - PubMed
-
- Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
