UNDR ROVER - a fast and accurate variant caller for targeted DNA sequencing
- PMID: 27083325
- PMCID: PMC4833922
- DOI: 10.1186/s12859-016-1014-9
UNDR ROVER - a fast and accurate variant caller for targeted DNA sequencing
Abstract
Background: Previously, we described ROVER, a DNA variant caller which identifies genetic variants from PCR-targeted massively parallel sequencing (MPS) datasets generated by the Hi-Plex protocol. ROVER permits stringent filtering of sequencing chemistry-induced errors by requiring reported variants to appear in both reads of overlapping pairs above certain thresholds of occurrence. ROVER was developed in tandem with Hi-Plex and has been used successfully to screen for genetic mutations in the breast cancer predisposition gene PALB2. ROVER is applied to MPS data in BAM format and, therefore, relies on sequence reads being mapped to a reference genome. In this paper, we describe an improvement to ROVER, called UNDR ROVER (Unmapped primer-Directed ROVER), which accepts MPS data in FASTQ format, avoiding the need for a computationally expensive mapping stage. It does so by taking advantage of the location-specific nature of PCR-targeted MPS data.
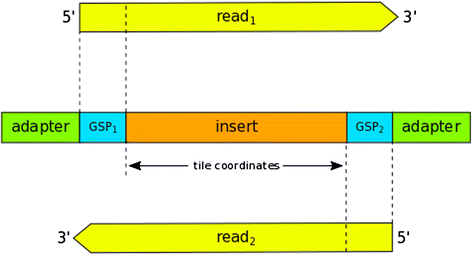
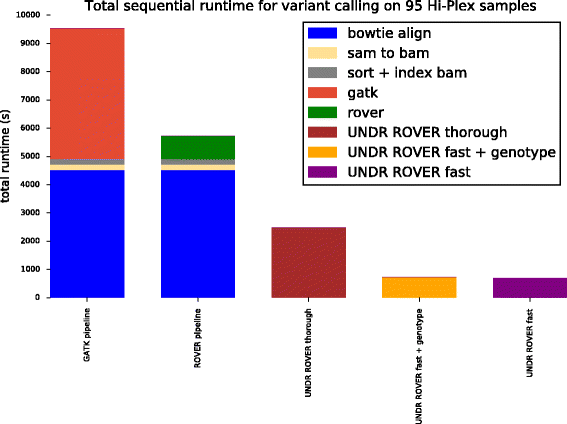
Results: The UNDR ROVER algorithm achieves the same stringent variant calling as its predecessor with a significant runtime performance improvement. In one indicative sequencing experiment, UNDR ROVER (in its fastest mode) required 8-fold less sequential computation time than the ROVER pipeline and 13-fold less sequential computation time than a variant calling pipeline based on the popular GATK tool. UNDR ROVER is implemented in Python and runs on all popular POSIX-like operating systems (Linux, OS X). It requires as input a tab-delimited format file containing primer sequence information, a FASTA format file containing the reference genome sequence, and paired FASTQ files containing sequence reads. Primer sequences at the 5' end of reads associate read-pairs with their targeted amplicon and, thus, their expected corresponding coordinates in the reference genome. The primer-intervening sequence of each read is compared against the reference sequence from the same location and variants are identified using the same algorithm as ROVER. Specifically, for a variant to be 'called' it must appear at the same location in both of the overlapping reads above user-defined thresholds of minimum number of reads and proportion of reads.
Conclusions: UNDR ROVER provides the same rapid and accurate genetic variant calling as its predecessor with greatly reduced computational costs.
Keywords: Hi-Plex; Massively parallel sequencing; PCR-MPS; ROVER; Targeted sequencing; Variant calling.
Figures

References
-
- Nguyen-Dumont T, Hammet F, Mahmoodi M, Pope BJ, Giles GG, Hopper GG, Southey MC, Park DJ. Abridged adapter primers increase the target scope of Hi-Plex. Biotechniques. 2014;58(1):33–36. - PubMed
-
- Nguyen-Dumont T, Hammet F, Mahmoodi M, Tsimiklis H, Teo ZL, Li R, Pope BJ, Terry MB, Buys SS, Daly M, et al. Mutation screening of PALB2 in clinically ascertained families from the Breast Cancer Family Registry. Breast Cancer Res Treat. 2015;149(2):547–554. doi: 10.1007/s10549-014-3260-8. - DOI - PMC - PubMed
-
- Sequence Alignment/Map Format Specification, Version 1. [http://samtools.github.io/hts-specs/SAMv1.pdf]. Accessed 14 Apr 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

