CTCF: making the right connections
- PMID: 27083996
- PMCID: PMC4840295
- DOI: 10.1101/gad.277863.116
CTCF: making the right connections
Abstract
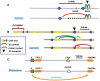
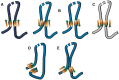
The role of the zinc finger protein CTCF in organizing the genome within the nucleus is now well established. Widely separated sites on DNA, occupied by both CTCF and the cohesin complex, make physical contacts that create large loop domains. Additional contacts between loci within those domains, often also mediated by CTCF, tend to be favored over contacts between loci in different domains. A large number of studies during the past 2 years have addressed the questions: How are these loops generated? What are the effects of disrupting them? Are there rules governing large-scale genome organization? It now appears that the strongest and evolutionarily most conserved of these CTCF interactions have specific rules for the orientation of the paired CTCF sites, implying the existence of a nonequilibrium mechanism of generation. Recent experiments that invert, delete, or inactivate one of a mating CTCF pair result in major changes in patterns of organization and gene expression in the surrounding regions. What remain to be determined are the detailed molecular mechanisms for re-establishing loop domains and maintaining them after replication and mitosis. As recently published data show, some mechanisms may involve interactions with noncoding RNAs as well as protein cofactors. Many CTCF sites are also involved in other functions such as modulation of RNA splicing and specific regulation of gene expression, and the relationship between these activities and loop formation is another unanswered question that should keep investigators occupied for some time.
Keywords: chromatin; insulators; topologically associated domains.
Published by Cold Spring Harbor Laboratory Press.
Figures

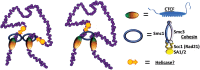

References
-
- Arib G, Cleard F, Maeda RK, Karch F. 2015. Following the intracellular localization of the iab-8ncRNA of the bithorax complex using the MS2-MCP-GFP system. Mech Dev 138(Pt 2): 133–140. - PubMed
-
- Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
