Tetratricopeptide repeat factor XAB2 mediates the end resection step of homologous recombination
- PMID: 27084940
- PMCID: PMC4937314
- DOI: 10.1093/nar/gkw275
Tetratricopeptide repeat factor XAB2 mediates the end resection step of homologous recombination
Abstract
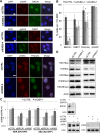
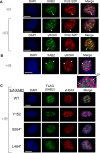
We examined the influence of the tetratricopeptide repeat factor XAB2 on chromosomal break repair, and found that XAB2 promotes end resection that generates the 3' ssDNA intermediate for homologous recombination (HR). Namely, XAB2 is important for chromosomal double-strand break (DSB) repair via two pathways of HR that require end resection as an intermediate step, end resection of camptothecin (Cpt)-induced DNA damage, and RAD51 recruitment to ionizing radiation induced foci (IRIF), which requires end resection. Furthermore, XAB2 mediates specific aspects of the DNA damage response associated with end resection proficiency: CtIP hyperphosphorylation induced by Cpt and BRCA1 IRIF. XAB2 also promotes histone acetylation events linked to HR proficiency. From truncation mutation analysis, the capacity for XAB2 to promote HR correlates with its ability to form a complex with ISY1 and PRP19, which show a similar influence as XAB2 on HR. This XAB2 complex localizes to punctate structures consistent with interchromatin granules that show a striking adjacent-localization to the DSB marker γH2AX. In summary, we suggest that the XAB2 complex mediates DNA damage response events important for the end resection step of HR, and speculate that its adjacent-localization relative to DSBs marked by γH2AX is important for this function.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

