From selection hits to clinical leads: progress in aptamer discovery
- PMID: 27088106
- PMCID: PMC4822646
- DOI: 10.1038/mtm.2016.14
From selection hits to clinical leads: progress in aptamer discovery
Abstract
Aptamers were discovered more than 25 years ago, yet only one has been approved by the US Food and Drug Administration to date. With some noteworthy advances in their chemical design and the enzymes we use to make them, aptamers and aptamer-based therapeutics have seen a resurgence in interest. New aptamer drugs are being approved for clinical evaluation, and it is certain that we will see increasingly more aptamers and aptamer-like drugs in the future. In this review, we will discuss the production of aptamers with an emphasis on the advances and modifications that enabled early aptamers to succeed in clinical trials as well as those that are likely to be important for future generations of these drugs.
Figures
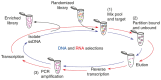
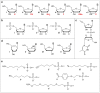
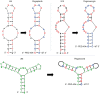
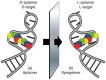
References
-
- Tuerk, C and Gold, L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. - PubMed
-
- Ellington, AD and Szostak, JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. - PubMed
-
- Hall, B, Arshad, S, Seo, K, Bowman, C, Corley, M, Jhaveri, SD et al. (2009). In Vitro Selection of RNA Aptamers to a Protein Target by Filter Immobilization. John Wiley & SonsHoboken. pp. 24.3.1–24.3.27. - PubMed
-
- Yan, A and Levy, M (2014). Cell internalization SELEX: in vitro selection for molecules that internalize into cells. Methods Mol Biol 1103: 241–265. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

