Ndfip-mediated degradation of Jak1 tunes cytokine signalling to limit expansion of CD4+ effector T cells
- PMID: 27088444
- PMCID: PMC4837450
- DOI: 10.1038/ncomms11226
Ndfip-mediated degradation of Jak1 tunes cytokine signalling to limit expansion of CD4+ effector T cells
Abstract
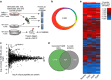
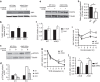
Nedd4 family E3 ubiquitin ligases have been shown to restrict T-cell function and impact T-cell differentiation. We show here that Ndfip1 and Ndfip2, activators of Nedd4 family ligases, together limit accumulation and function of effector CD4+ T cells. Using a three-part proteomics approach in primary T cells, we identify stabilization of Jak1 in Ndfip1/2-deficient T cells stimulated through the TCR. Jak1 degradation is aborted in activated T cells that lack Ndfips. In wild-type cells, Jak1 degradation lessens CD4+ cell sensitivity to cytokines during TCR stimulation, while in Ndfip-deficient cells cytokine responsiveness persists, promoting increased expansion and survival of pathogenic effector T cells. Thus, Ndfip1/Ndfip2 regulate the cross talk between the T-cell receptor and cytokine signalling pathways to limit inappropriate T-cell responses.
Figures






References
-
- Fang D. et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3, 281–287 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

