The Atlantic salmon genome provides insights into rediploidization
- PMID: 27088604
- PMCID: PMC8127823
- DOI: 10.1038/nature17164
The Atlantic salmon genome provides insights into rediploidization
Abstract
The whole-genome duplication 80 million years ago of the common ancestor of salmonids (salmonid-specific fourth vertebrate whole-genome duplication, Ss4R) provides unique opportunities to learn about the evolutionary fate of a duplicated vertebrate genome in 70 extant lineages. Here we present a high-quality genome assembly for Atlantic salmon (Salmo salar), and show that large genomic reorganizations, coinciding with bursts of transposon-mediated repeat expansions, were crucial for the post-Ss4R rediploidization process. Comparisons of duplicate gene expression patterns across a wide range of tissues with orthologous genes from a pre-Ss4R outgroup unexpectedly demonstrate far more instances of neofunctionalization than subfunctionalization. Surprisingly, we find that genes that were retained as duplicates after the teleost-specific whole-genome duplication 320 million years ago were not more likely to be retained after the Ss4R, and that the duplicate retention was not influenced to a great extent by the nature of the predicted protein interactions of the gene products. Finally, we demonstrate that the Atlantic salmon assembly can serve as a reference sequence for the study of other salmonids for a range of purposes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

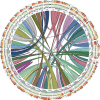
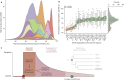






Comment in
-
Subfunctionalization versus neofunctionalization after whole-genome duplication.Nat Genet. 2018 Jul;50(7):908-909. doi: 10.1038/s41588-018-0162-4. Nat Genet. 2018. PMID: 29955176 No abstract available.
References
-
- Nelson, J. S. Fishes of the World (John Wiley & Sons, 2006)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

