Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells
- PMID: 27090285
- PMCID: PMC4878613
- DOI: 10.1261/rna.053561.115
Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells
Abstract
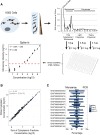
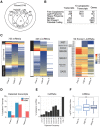
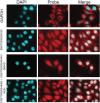
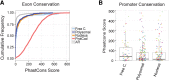
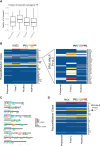
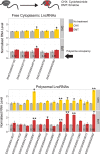
Recent footprinting studies have made the surprising observation that long noncoding RNAs (lncRNAs) physically interact with ribosomes. However, these findings remain controversial, and the overall proportion of cytoplasmic lncRNAs involved is unknown. Here we make a global, absolute estimate of the cytoplasmic and ribosome-associated population of stringently filtered lncRNAs in a human cell line using polysome profiling coupled to spike-in normalized microarray analysis. Fifty-four percent of expressed lncRNAs are detected in the cytoplasm. The majority of these (70%) have >50% of their cytoplasmic copies associated with polysomal fractions. These interactions are lost upon disruption of ribosomes by puromycin. Polysomal lncRNAs are distinguished by a number of 5' mRNA-like features, including capping and 5'UTR length. On the other hand, nonpolysomal "free cytoplasmic" lncRNAs have more conserved promoters and a wider range of expression across cell types. Exons of polysomal lncRNAs are depleted of endogenous retroviral insertions, suggesting a role for repetitive elements in lncRNA localization. Finally, we show that blocking of ribosomal elongation results in stabilization of many associated lncRNAs. Together these findings suggest that the ribosome is the default destination for the majority of cytoplasmic long noncoding RNAs and may play a role in their degradation.
Keywords: cytoplasm; degradation; long noncoding RNA; ribosome; ribosome profiling; translation; transposable element.
© 2016 Carlevaro-Fita et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Beilharz TH, Preiss T. 2004. Translational profiling: the genome-wide measure of the nascent proteome. Brief Funct Genomic Proteomic 3: 103–111. - PubMed
-
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
