Dynamic and flexible H3K9me3 bridging via HP1β dimerization establishes a plastic state of condensed chromatin
- PMID: 27090491
- PMCID: PMC4838890
- DOI: 10.1038/ncomms11310
Dynamic and flexible H3K9me3 bridging via HP1β dimerization establishes a plastic state of condensed chromatin
Abstract
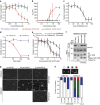
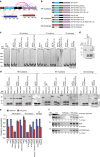
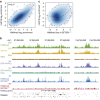
Histone H3 trimethylation of lysine 9 (H3K9me3) and proteins of the heterochromatin protein 1 (HP1) family are hallmarks of heterochromatin, a state of compacted DNA essential for genome stability and long-term transcriptional silencing. The mechanisms by which H3K9me3 and HP1 contribute to chromatin condensation have been speculative and controversial. Here we demonstrate that human HP1β is a prototypic HP1 protein exemplifying most basal chromatin binding and effects. These are caused by dimeric and dynamic interaction with highly enriched H3K9me3 and are modulated by various electrostatic interfaces. HP1β bridges condensed chromatin, which we postulate stabilizes the compacted state. In agreement, HP1β genome-wide localization follows H3K9me3-enrichment and artificial bridging of chromatin fibres is sufficient for maintaining cellular heterochromatic conformation. Overall, our findings define a fundamental mechanism for chromatin higher order structural changes caused by HP1 proteins, which might contribute to the plastic nature of condensed chromatin.
Figures





References
-
- Grewal S. I. & Jia S. Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 (2007). - PubMed
-
- Maison C. & Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 (2004). - PubMed
-
- Fodor B. D., Shukeir N., Reuter G. & Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu. Rev. Cell Dev. Biol. 26, 471–501 (2010). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

