Nuclear RNA-seq of single neurons reveals molecular signatures of activation
- PMID: 27090946
- PMCID: PMC4838832
- DOI: 10.1038/ncomms11022
Nuclear RNA-seq of single neurons reveals molecular signatures of activation
Erratum in
-
Corrigendum: Nuclear RNA-seq of single neurons reveals molecular signatures of activation.Nat Commun. 2016 Jun 14;7:12020. doi: 10.1038/ncomms12020. Nat Commun. 2016. PMID: 27295945 Free PMC article. No abstract available.
-
Corrigendum: Nuclear RNA-seq of single neurons reveals molecular signatures of activation.Nat Commun. 2017 Mar 17;8:15047. doi: 10.1038/ncomms15047. Nat Commun. 2017. PMID: 28303884 Free PMC article. No abstract available.
Abstract
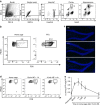
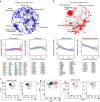
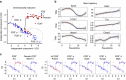
Single-cell sequencing methods have emerged as powerful tools for identification of heterogeneous cell types within defined brain regions. Application of single-cell techniques to study the transcriptome of activated neurons can offer insight into molecular dynamics associated with differential neuronal responses to a given experience. Through evaluation of common whole-cell and single-nuclei RNA-sequencing (snRNA-seq) methods, here we show that snRNA-seq faithfully recapitulates transcriptional patterns associated with experience-driven induction of activity, including immediate early genes (IEGs) such as Fos, Arc and Egr1. SnRNA-seq of mouse dentate granule cells reveals large-scale changes in the activated neuronal transcriptome after brief novel environment exposure, including induction of MAPK pathway genes. In addition, we observe a continuum of activation states, revealing a pseudotemporal pattern of activation from gene expression alone. In summary, snRNA-seq of activated neurons enables the examination of gene expression beyond IEGs, allowing for novel insights into neuronal activation patterns in vivo.
Conflict of interest statement
B.L. is currently an employee at Fluidigm. The other authors declare no competing financial interests.
Figures


References
-
- Hashimshony T., Wagner F., Sher N. & Yanai I. CEL-seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012). - PubMed
-
- Picelli S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc.. 9, 171–181 (2014). - PubMed
-
- Tang F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

