Supercomplex-associated Cox26 protein binds to cytochrome c oxidase
- PMID: 27091403
- PMCID: PMC7140176
- DOI: 10.1016/j.bbamcr.2016.04.012
Supercomplex-associated Cox26 protein binds to cytochrome c oxidase
Abstract
Here we identified a hydrophobic 6.4kDa protein, Cox26, as a novel component of yeast mitochondrial supercomplex comprising respiratory complexes III and IV. Multi-dimensional native and denaturing electrophoretic techniques were used to identify proteins interacting with Cox26. The majority of the Cox26 protein was found non-covalently bound to the complex IV moiety of the III-IV supercomplexes. A population of Cox26 was observed to exist in a disulfide bond partnership with the Cox2 subunit of complex IV. No pronounced growth phenotype for Cox26 deficiency was observed, indicating that Cox26 may not play a critical role in the COX enzymology, and we speculate that Cox26 may serve to regulate or support the Cox2 protein. Respiratory supercomplexes are assembled in the absence of the Cox26 protein, however their pattern slightly differs to the wild type III-IV supercomplex appearance. The catalytic activities of complexes III and IV were observed to be normal and respiration was comparable to wild type as long as cells were cultivated under normal growth conditions. Stress conditions, such as elevated temperatures resulted in mild decrease of respiration in non-fermentative media when the Cox26 protein was absent.
Keywords: Blue native electrophoresis; Cytochrome c oxidase; III–IV supercomplexes; Protein composition; Respiratory supercomplexes; Saccharomyces cerevisiae.
Copyright © 2016 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest
All authors have declared no conflicts of interest concerning the contents of the article.
Figures

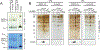
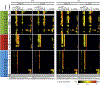
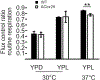

References
-
- Hatefi Y, The mitochondrial electron transport and oxidative phosphorylation system, Annu. Rev. Biochem 54 (1985) 1015–1069. - PubMed
-
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA, The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria, J. Biol. Chem 275 (2000) 18093–18098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

