Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif
- PMID: 27091468
- PMCID: PMC4884534
- DOI: 10.1016/j.jmb.2016.04.009
Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif
Abstract
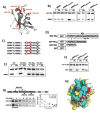
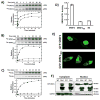
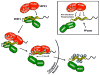
Multisite phosphorylation is required for the biological function of serine-arginine (SR) proteins, a family of essential regulators of mRNA splicing. These modifications are catalyzed by serine-arginine protein kinases (SRPKs) that phosphorylate numerous serines in arginine-serine-rich (RS) domains of SR proteins using a directional, C-to-N-terminal mechanism. The present studies explore how SRPKs govern this highly biased phosphorylation reaction and investigate biological roles of the observed directional phosphorylation mechanism. Using NMR spectroscopy with two separately expressed domains of SRSF1, we showed that several residues in the RNA-binding motif 2 interact with the N-terminal region of the RS domain (RS1). These contacts provide a structural framework that balances the activities of SRPK1 and the protein phosphatase PP1, thereby regulating the phosphoryl content of the RS domain. Disruption of the implicated intramolecular RNA-binding motif 2-RS domain interaction impairs both the directional phosphorylation mechanism and the nuclear translocation of SRSF1 demonstrating that the intrinsic phosphorylation bias is obligatory for SR protein biological function.
Keywords: NMR; RS domain; SR protein; kinetics; phosphorylation.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Conserved proline-directed phosphorylation regulates SR protein conformation and splicing function.Biochem J. 2015 Mar 1;466(2):311-22. doi: 10.1042/BJ20141373. Biochem J. 2015. PMID: 25529026 Free PMC article.
-
N-terminus of the protein kinase CLK1 induces SR protein hyperphosphorylation.Biochem J. 2014 Aug 15;462(1):143-52. doi: 10.1042/BJ20140494. Biochem J. 2014. PMID: 24869919 Free PMC article.
-
Splicing kinase SRPK1 conforms to the landscape of its SR protein substrate.Biochemistry. 2013 Oct 29;52(43):7595-605. doi: 10.1021/bi4010864. Epub 2013 Oct 15. Biochemistry. 2013. PMID: 24074032 Free PMC article.
-
Phosphorylation mechanism and structure of serine-arginine protein kinases.FEBS J. 2011 Feb;278(4):587-97. doi: 10.1111/j.1742-4658.2010.07992.x. Epub 2011 Jan 12. FEBS J. 2011. PMID: 21205204 Free PMC article. Review.
-
Serine-arginine protein kinases and their targets in viral infection and their inhibition.Cell Mol Life Sci. 2023 May 17;80(6):153. doi: 10.1007/s00018-023-04808-6. Cell Mol Life Sci. 2023. PMID: 37198350 Free PMC article. Review.
Cited by
-
Molecular Mechanism of SR Protein Kinase 1 Inhibition by the Herpes Virus Protein ICP27.mBio. 2019 Oct 22;10(5):e02551-19. doi: 10.1128/mBio.02551-19. mBio. 2019. PMID: 31641093 Free PMC article.
-
Multiple Phosphorylations of SR Protein SRSF3 and Its Binding to m6A Reader YTHDC1 in Human Cells.Cells. 2022 Apr 26;11(9):1461. doi: 10.3390/cells11091461. Cells. 2022. PMID: 35563766 Free PMC article.
-
Alternative Splicing in Angiogenesis.Int J Mol Sci. 2019 Apr 26;20(9):2067. doi: 10.3390/ijms20092067. Int J Mol Sci. 2019. PMID: 31027366 Free PMC article. Review.
-
SR Protein Kinases Regulate the Splicing of Cardiomyopathy-Relevant Genes via Phosphorylation of the RSRSP Stretch in RBM20.Genes (Basel). 2022 Aug 25;13(9):1526. doi: 10.3390/genes13091526. Genes (Basel). 2022. PMID: 36140694 Free PMC article.
-
RBM20 phosphorylation and its role in nucleocytoplasmic transport and cardiac pathogenesis.FASEB J. 2022 May;36(5):e22302. doi: 10.1096/fj.202101811RR. FASEB J. 2022. PMID: 35394688 Free PMC article.
References
-
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7. - PubMed
-
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–24. - PubMed
-
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–70. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

