Tissue-specific DNA demethylation is required for proper B-cell differentiation and function
- PMID: 27091986
- PMCID: PMC4983829
- DOI: 10.1073/pnas.1604365113
Tissue-specific DNA demethylation is required for proper B-cell differentiation and function
Abstract
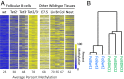
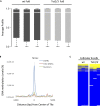
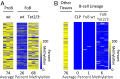
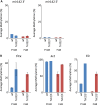
There is ample evidence that somatic cell differentiation during development is accompanied by extensive DNA demethylation of specific sites that vary between cell types. Although the mechanism of this process has not yet been elucidated, it is likely to involve the conversion of 5mC to 5hmC by Tet enzymes. We show that a Tet2/Tet3 conditional knockout at early stages of B-cell development largely prevents lineage-specific programmed demethylation events. This lack of demethylation affects the expression of nearby B-cell lineage genes by impairing enhancer activity, thus causing defects in B-cell differentiation and function. Thus, tissue-specific DNA demethylation appears to be necessary for proper somatic cell development in vivo.
Keywords: DMRs; Tet2/Tet3; chromatin; differentially methylated regions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
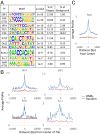



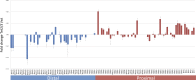
References
-
- Guo F, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–458. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

