Gene expression profiling of microbial activities and interactions in sediments under haloclines of E. Mediterranean deep hypersaline anoxic basins
- PMID: 27093045
- PMCID: PMC5113848
- DOI: 10.1038/ismej.2016.58
Gene expression profiling of microbial activities and interactions in sediments under haloclines of E. Mediterranean deep hypersaline anoxic basins
Abstract
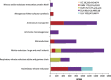
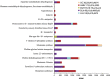
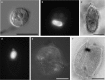
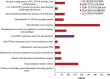
Deep-sea hypersaline anoxic basins (DHABs) in the Eastern Mediterranean Sea are considered some of the most polyextreme habitats on Earth. In comparison to microbial activities occurring within the haloclines and brines of these unusual water column habitats near the Mediterranean seafloor, relatively little is known about microbial metabolic activities in the underlying sediments. In addition, it is not known whether activities are shaped by the unique chemistries of the different DHAB brines and whether evidence exists for active microbial eukaryotes in those sediments. Metatranscriptome analysis was applied to sediment samples collected using ROV Jason from underneath the haloclines of Urania, Discovery and L'Atalante DHABs and a control site. We report on expression of genes associated with sulfur and nitrogen cycling, putative osmolyte biosynthetic pathways and ion transporters, trace metal detoxification, selected eukaryotic activities (particularly of fungi), microbe-microbe interactions, and motility in sediments underlying the haloclines of three DHABs. Relative to our control sediment sample collected outside of Urania Basin, microbial communities (including eukaryotes) in the Urania and Discovery DHAB sediments showed upregulation of expressed genes associated with nitrogen transformations, osmolyte biosynthesis, heavy metals resistance and metabolism, eukaryotic organelle functions, and cell-cell interactions. Sediments underlying DHAB haloclines that have cumulative physico-chemical stressors within the limits of tolerance for microoorganisms can therefore be hotspots of activity in the deep Mediterranean Sea.
Figures


Similar articles
-
Metazoans of redoxcline sediments in Mediterranean deep-sea hypersaline anoxic basins.BMC Biol. 2015 Dec 10;13:105. doi: 10.1186/s12915-015-0213-6. BMC Biol. 2015. PMID: 26652623 Free PMC article.
-
Inter-comparison of the potentially active prokaryotic communities in the halocline sediments of Mediterranean deep-sea hypersaline basins.Extremophiles. 2015 Sep;19(5):949-60. doi: 10.1007/s00792-015-0770-1. Epub 2015 Jul 16. Extremophiles. 2015. PMID: 26174531
-
Benthic protists and fungi of Mediterranean deep hypsersaline anoxic basin redoxcline sediments.Front Microbiol. 2014 Nov 12;5:605. doi: 10.3389/fmicb.2014.00605. eCollection 2014. Front Microbiol. 2014. PMID: 25452749 Free PMC article.
-
Microbial ecology of deep-sea hypersaline anoxic basins.FEMS Microbiol Ecol. 2018 Jul 1;94(7). doi: 10.1093/femsec/fiy085. FEMS Microbiol Ecol. 2018. PMID: 29905791 Review.
-
Current state of athalassohaline deep-sea hypersaline anoxic basin research-recommendations for future work and relevance to astrobiology.Environ Microbiol. 2021 Jul;23(7):3360-3369. doi: 10.1111/1462-2920.15414. Epub 2021 Feb 15. Environ Microbiol. 2021. PMID: 33538392 Review.
Cited by
-
Current Insight into Traditional and Modern Methods in Fungal Diversity Estimates.J Fungi (Basel). 2022 Feb 24;8(3):226. doi: 10.3390/jof8030226. J Fungi (Basel). 2022. PMID: 35330228 Free PMC article. Review.
-
Insights into fungal diversity of a shallow-water hydrothermal vent field at Kueishan Island, Taiwan by culture-based and metabarcoding analyses.PLoS One. 2019 Dec 30;14(12):e0226616. doi: 10.1371/journal.pone.0226616. eCollection 2019. PLoS One. 2019. PMID: 31887170 Free PMC article.
-
Life on the edge: active microbial communities in the Kryos MgCl2-brine basin at very low water activity.ISME J. 2018 Jun;12(6):1414-1426. doi: 10.1038/s41396-018-0107-z. Epub 2018 Apr 17. ISME J. 2018. PMID: 29666446 Free PMC article.
-
Fungi in the Marine Environment: Open Questions and Unsolved Problems.mBio. 2019 Mar 5;10(2):e01189-18. doi: 10.1128/mBio.01189-18. mBio. 2019. PMID: 30837337 Free PMC article. Review.
-
Multiomic analysis of different horse breeds reveals that gut microbial butyrate enhances racehorse athletic performance.NPJ Biofilms Microbiomes. 2025 May 24;11(1):87. doi: 10.1038/s41522-025-00730-w. NPJ Biofilms Microbiomes. 2025. PMID: 40410196 Free PMC article.
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. (2002). How cells regulate their cytoskeletal filaments. In: Molecular Biology of the Cell 4th edition Garland Science: New York, USA.
-
- Alexander E, Stock A, Breiner HW, Behnke A, Bunge J, Yakimov MM et al. (2009). Microbial eukaryotes in the hypersaline anoxic L'Atalante deep-sea basin. Env Microbiol 11: 360–381. - PubMed
-
- Bay DC, Rommens KL, Turner RJ. (2008). Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta 1778: 1814–1838. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

