Genomics of a phototrophic nitrite oxidizer: insights into the evolution of photosynthesis and nitrification
- PMID: 27093047
- PMCID: PMC5113846
- DOI: 10.1038/ismej.2016.56
Genomics of a phototrophic nitrite oxidizer: insights into the evolution of photosynthesis and nitrification
Abstract
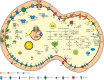
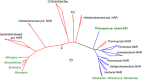
Oxygenic photosynthesis evolved from anoxygenic ancestors before the rise of oxygen ~2.32 billion years ago; however, little is known about this transition. A high redox potential reaction center is a prerequisite for the evolution of the water-oxidizing complex of photosystem II. Therefore, it is likely that high-potential phototrophy originally evolved to oxidize alternative electron donors that utilized simpler redox chemistry, such as nitrite or Mn. To determine whether nitrite could have had a role in the transition to high-potential phototrophy, we sequenced and analyzed the genome of Thiocapsa KS1, a Gammaproteobacteria capable of anoxygenic phototrophic nitrite oxidation. The genome revealed a high metabolic flexibility, which likely allows Thiocapsa KS1 to colonize a great variety of habitats and to persist under fluctuating environmental conditions. We demonstrate that Thiocapsa KS1 does not utilize a high-potential reaction center for phototrophic nitrite oxidation, which suggests that this type of phototrophic nitrite oxidation did not drive the evolution of high-potential phototrophy. In addition, phylogenetic and biochemical analyses of the nitrite oxidoreductase (NXR) from Thiocapsa KS1 illuminate a complex evolutionary history of nitrite oxidation. Our results indicate that the NXR in Thiocapsa originates from a different nitrate reductase clade than the NXRs in chemolithotrophic nitrite oxidizers, suggesting that multiple evolutionary trajectories led to modern nitrite-oxidizing bacteria.
Figures

References
-
- Allen JP, Williams JC. (2014). Energetics of cofactors in photosynthetic complexes: relationship between protein–cofactor interactions and midpoint potentials. In: Golbeck JH, Van der Est A (eds) The Biophysics of Photosynthesis. Springer: New York, NY, USA, pp 275–295.
-
- Allen JP, Williams JC. (2011). The evolutionary pathway from anoxygenic to oxygenic photosynthesis examined by comparison of the properties of photosystem II and bacterial reaction centers. Photosyn Res 107: 59–69. - PubMed
-
- Blankenship RE. (2014) Molecular Mechanisms of Photosynthesis, 2nd edn. Wiley-Blackwell.
-
- Blasco F, Santos Dos JP, Magalon A, Frixon C, Guigliarelli B, Santini CL et al. (1998). NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol Microbiol 28: 435–447. - PubMed
-
- Bock E. (1976). Growth of Nitrobacter in the presence of organic matter. Arch Microbiol 108: 305–312. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

