Polo Kinase Phosphorylates Miro to Control ER-Mitochondria Contact Sites and Mitochondrial Ca(2+) Homeostasis in Neural Stem Cell Development
- PMID: 27093086
- PMCID: PMC4839004
- DOI: 10.1016/j.devcel.2016.03.023
Polo Kinase Phosphorylates Miro to Control ER-Mitochondria Contact Sites and Mitochondrial Ca(2+) Homeostasis in Neural Stem Cell Development
Abstract
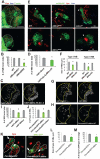
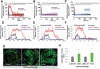
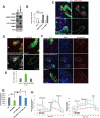
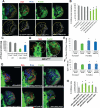
Mitochondria play central roles in buffering intracellular Ca²⁺ transients. While basal mitochondrial Ca²⁺ (Ca²⁺ mito) is needed to maintain organellar physiology, Ca²⁺ mito overload can lead to cell death. How Ca²⁺ mito homeostasis is regulated is not well understood. Here we show that Miro, a known component of the mitochondrial transport machinery, regulates Drosophila neural stem cell (NSC) development through Ca²⁺ mito homeostasis control, independent of its role in mitochondrial transport. Miro interacts with Ca²⁺ transporters at the ER-mitochondria contact site (ERMCS). Its inactivation causes Ca²⁺ mito depletion and metabolic impairment, whereas its overexpression results in Ca²⁺ mito overload, mitochondrial morphology change, and apoptotic response. Both conditions impaired NSC lineage progression. Ca²⁺ mito homeostasis is influenced by Polo-mediated phosphorylation of a conserved residue in Miro, which positively regulates Miro localization to, and the integrity of, ERMCS. Our results elucidate a regulatory mechanism underlying Ca²⁺ mito homeostasis and how its dysregulation may affect NSC metabolism/development and contribute to disease.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
References
-
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
