The neurogenetics of alternative splicing
- PMID: 27094079
- PMCID: PMC4861142
- DOI: 10.1038/nrn.2016.27
The neurogenetics of alternative splicing
Abstract
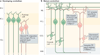
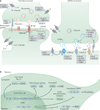
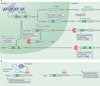
Alternative precursor-mRNA splicing is a key mechanism for regulating gene expression in mammals and is controlled by specialized RNA-binding proteins. The misregulation of splicing is implicated in multiple neurological disorders. We describe recent mouse genetic studies of alternative splicing that reveal its critical role in both neuronal development and the function of mature neurons. We discuss the challenges in understanding the extensive genetic programmes controlled by proteins that regulate splicing, both during development and in the adult brain.
Figures
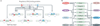

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

