A somatic reference standard for cancer genome sequencing
- PMID: 27094764
- PMCID: PMC4837349
- DOI: 10.1038/srep24607
A somatic reference standard for cancer genome sequencing
Abstract
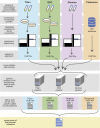
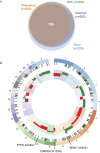
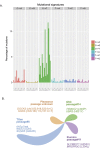
Large-scale multiplexed identification of somatic alterations in cancer has become feasible with next generation sequencing (NGS). However, calibration of NGS somatic analysis tools has been hampered by a lack of tumor/normal reference standards. We thus performed paired PCR-free whole genome sequencing of a matched metastatic melanoma cell line (COLO829) and normal across three lineages and across separate institutions, with independent library preparations, sequencing, and analysis. We generated mean mapped coverages of 99X for COLO829 and 103X for the paired normal across three institutions. Results were combined with previously generated data allowing for comparison to a fourth lineage on earlier NGS technology. Aggregate variant detection led to the identification of consensus variants, including key events that represent hallmark mutation types including amplified BRAF V600E, a CDK2NA small deletion, a 12 kb PTEN deletion, and a dinucleotide TERT promoter substitution. Overall, common events include >35,000 point mutations, 446 small insertion/deletions, and >6,000 genes affected by copy number changes. We present this reference to the community as an initial standard for enabling quantitative evaluation of somatic mutation pipelines across institutions.
Figures
References
-
- Zook J. M. et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 32, 246–251 (2014). - PubMed
-
- Eberle M. A. et al. Platinum Genomes: A systematic assessment of variant accuracy using a large family pedigree. Presented at the 60th Annual Meeting of The American Society of Human Genetics, October 22–26, Boston (2013)
-
- Simen B. B. et al. Validation of a Next-Generation-Sequencing Cancer Panel for Use in the Clinical Laboratory. Arch Pathol Lab Med. 139, 508–517 (2015). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

