Packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA depends upon conserved long-range interactions (LRIs) between U5 and gag sequences
- PMID: 27095024
- PMCID: PMC4878616
- DOI: 10.1261/rna.055731.115
Packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA depends upon conserved long-range interactions (LRIs) between U5 and gag sequences
Abstract
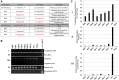
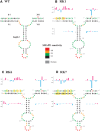
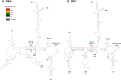
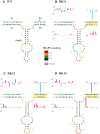
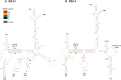
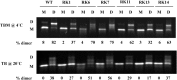
MPMV has great potential for development as a vector for gene therapy. In this respect, precisely defining the sequences and structural motifs that are important for dimerization and packaging of its genomic RNA (gRNA) are of utmost importance. A distinguishing feature of the MPMV gRNA packaging signal is two phylogenetically conserved long-range interactions (LRIs) between U5 and gag complementary sequences, LRI-I and LRI-II. To test their biological significance in the MPMV life cycle, we introduced mutations into these structural motifs and tested their effects on MPMV gRNA packaging and propagation. Furthermore, we probed the structure of key mutants using SHAPE (selective 2'hydroxyl acylation analyzed by primer extension). Disrupting base-pairing of the LRIs affected gRNA packaging and propagation, demonstrating their significance to the MPMV life cycle. A double mutant restoring a heterologous LRI-I was fully functional, whereas a similar LRI-II mutant failed to restore gRNA packaging and propagation. These results demonstrate that while LRI-I acts at the structural level, maintaining base-pairing is not sufficient for LRI-II function. In addition, in vitro RNA dimerization assays indicated that the loss of RNA packaging in LRI mutants could not be attributed to the defects in dimerization. Our findings suggest that U5-gag LRIs play an important architectural role in maintaining the structure of the 5' region of the MPMV gRNA, expanding the crucial role of LRIs to the nonlentiviral group of retroviruses.
Keywords: Mason-Pfizer monkey virus (MPMV); RNA packaging and dimerization; RNA secondary structure; SHAPE (selective 2′hydroxyl acylation analyzed by primer extension); long-range interactions (LRI); retroviruses.
© 2016 Kalloush et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


Similar articles
-
Stabilizing role of structural elements within the 5´ Untranslated Region (UTR) and gag sequences in Mason-Pfizer monkey virus (MPMV) genomic RNA packaging.RNA Biol. 2019 May;16(5):612-625. doi: 10.1080/15476286.2019.1572424. Epub 2019 Feb 17. RNA Biol. 2019. PMID: 30773097 Free PMC article.
-
SHAPE analysis of the 5' end of the Mason-Pfizer monkey virus (MPMV) genomic RNA reveals structural elements required for genome dimerization.RNA. 2013 Dec;19(12):1648-58. doi: 10.1261/rna.040931.113. Epub 2013 Oct 23. RNA. 2013. PMID: 24152551 Free PMC article.
-
Role of Purine-Rich Regions in Mason-Pfizer Monkey Virus (MPMV) Genomic RNA Packaging and Propagation.Front Microbiol. 2020 Nov 5;11:595410. doi: 10.3389/fmicb.2020.595410. eCollection 2020. Front Microbiol. 2020. PMID: 33250884 Free PMC article.
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
-
Retroviral RNA Dimerization: From Structure to Functions.Front Microbiol. 2018 Mar 22;9:527. doi: 10.3389/fmicb.2018.00527. eCollection 2018. Front Microbiol. 2018. PMID: 29623074 Free PMC article. Review.
Cited by
-
MMTV RNA packaging requires an extended long-range interaction for productive Gag binding to packaging signals.PLoS Biol. 2024 Oct 3;22(10):e3002827. doi: 10.1371/journal.pbio.3002827. eCollection 2024 Oct. PLoS Biol. 2024. PMID: 39361708 Free PMC article.
-
Rous Sarcoma Virus Genomic RNA Dimerization Capability In Vitro Is Not a Prerequisite for Viral Infectivity.Viruses. 2020 May 22;12(5):568. doi: 10.3390/v12050568. Viruses. 2020. PMID: 32455905 Free PMC article.
-
The Life-Cycle of the HIV-1 Gag-RNA Complex.Viruses. 2016 Sep 10;8(9):248. doi: 10.3390/v8090248. Viruses. 2016. PMID: 27626439 Free PMC article. Review.
-
Expression, purification, and characterization of biologically active full-length Mason-Pfizer monkey virus (MPMV) Pr78Gag.Sci Rep. 2018 Aug 7;8(1):11793. doi: 10.1038/s41598-018-30142-0. Sci Rep. 2018. PMID: 30087395 Free PMC article.
-
The bifurcated stem loop 4 (SL4) is crucial for efficient packaging of mouse mammary tumor virus (MMTV) genomic RNA.RNA Biol. 2018;15(8):1047-1059. doi: 10.1080/15476286.2018.1486661. Epub 2018 Jul 5. RNA Biol. 2018. PMID: 29929424 Free PMC article.
References
-
- Abbink TE, Berkhout B. 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem 278: 11601–11611. - PubMed
-
- Abd El-Wahab EW, Smyth RP, Mailler E, Bernacchi S, Vivet-Boudou V, Hijnen M, Jossinet F, Mak J, Paillart JC, Marquet R. 2014. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat Commun 5: 4304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
