iGEMS: an integrated model for identification of alternative exon usage events
- PMID: 27095197
- PMCID: PMC4914109
- DOI: 10.1093/nar/gkw263
iGEMS: an integrated model for identification of alternative exon usage events
Abstract
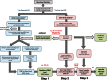
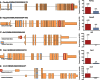
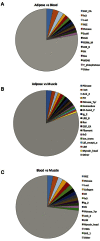
DNA microarrays and RNAseq are complementary methods for studying RNA molecules. Current computational methods to determine alternative exon usage (AEU) using such data require impractical visual inspection and still yield high false-positive rates. Integrated Gene and Exon Model of Splicing (iGEMS) adapts a gene-level residuals model with a gene size adjusted false discovery rate and exon-level analysis to circumvent these limitations. iGEMS was applied to two new DNA microarray datasets, including the high coverage Human Transcriptome Arrays 2.0 and performance was validated using RT-qPCR. First, AEU was studied in adipocytes treated with (n = 9) or without (n = 8) the anti-diabetes drug, rosiglitazone. iGEMS identified 555 genes with AEU, and robust verification by RT-qPCR (∼90%). Second, in a three-way human tissue comparison (muscle, adipose and blood, n = 41) iGEMS identified 4421 genes with at least one AEU event, with excellent RT-qPCR verification (95%, n = 22). Importantly, iGEMS identified a variety of AEU events, including 3'UTR extension, as well as exon inclusion/exclusion impacting on protein kinase and extracellular matrix domains. In conclusion, iGEMS is a robust method for identification of AEU while the variety of exon usage between human tissues is 5-10 times more prevalent than reported by the Genotype-Tissue Expression consortium using RNA sequencing.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- De Conti L., Baralle M., Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA. 2013;4:49–60. - PubMed
-
- Blencowe B.J., Ahmad S., Lee L.J. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. - PubMed
-
- Chen F.C. Are all of the human exons alternatively spliced? Brief. Bioinform. 2014;15:542–551. - PubMed
-
- Thorsen K., Sørensen K.D., Brems-Eskildsen A.S., Modin C., Gaustadnes M., Hein A.-M.K., Kruhøffer M., Laurberg S., Borre M., Wang K., et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol. Cell. Proteomics. 2008;7:1214–1224. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

