A multivariate nonlinear mixed effects model for longitudinal image analysis: Application to amyloid imaging
- PMID: 27095307
- PMCID: PMC4912927
- DOI: 10.1016/j.neuroimage.2016.04.001
A multivariate nonlinear mixed effects model for longitudinal image analysis: Application to amyloid imaging
Abstract
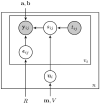
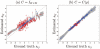
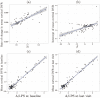
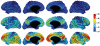
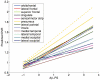
It is important to characterize the temporal trajectories of disease-related biomarkers in order to monitor progression and identify potential points of intervention. These are especially important for neurodegenerative diseases, as therapeutic intervention is most likely to be effective in the preclinical disease stages prior to significant neuronal damage. Neuroimaging allows for the measurement of structural, functional, and metabolic integrity of the brain at the level of voxels, whose volumes are on the order of mm(3). These voxelwise measurements provide a rich collection of disease indicators. Longitudinal neuroimaging studies enable the analysis of changes in these voxelwise measures. However, commonly used longitudinal analysis approaches, such as linear mixed effects models, do not account for the fact that individuals enter a study at various disease stages and progress at different rates, and generally consider each voxelwise measure independently. We propose a multivariate nonlinear mixed effects model for estimating the trajectories of voxelwise neuroimaging biomarkers from longitudinal data that accounts for such differences across individuals. The method involves the prediction of a progression score for each visit based on a collective analysis of voxelwise biomarker data within an expectation-maximization framework that efficiently handles large amounts of measurements and variable number of visits per individual, and accounts for spatial correlations among voxels. This score allows individuals with similar progressions to be aligned and analyzed together, which enables the construction of a trajectory of brain changes as a function of an underlying progression or disease stage. We apply our method to studying cortical β-amyloid deposition, a hallmark of preclinical Alzheimer's disease, as measured using positron emission tomography. Results on 104 individuals with a total of 300 visits suggest that precuneus is the earliest cortical region to accumulate amyloid, closely followed by the cingulate and frontal cortices, then by the lateral parietal cortex. The extracted progression scores reveal a pattern similar to mean cortical distribution volume ratio (DVR), an index of global brain amyloid levels. The proposed method can be applied to other types of longitudinal imaging data, including metabolism, blood flow, tau, and structural imaging-derived measures, to extract individualized summary scores indicating disease progression and to provide voxelwise trajectories that can be compared between brain regions.
Keywords: Amyloid imaging; Longitudinal image analysis; Progression score.
Published by Elsevier Inc.
Figures
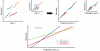


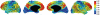


References
-
- Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, Marcus DS, Cairns NJ, Xie X, Blazey TM, Holtzman DM, Santacruz A, Buckles V, Oliver A, Moulder K, Aisen PS, Ghetti B, Klunk WE, McDade E, Martins RN, Masters CL, Mayeux R, Ringman JM, Rossor MN, Schofield PR, Sperling RA, Salloway S, Morris JC. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. New England Journal of Medicine. 2012;367:795–804. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

